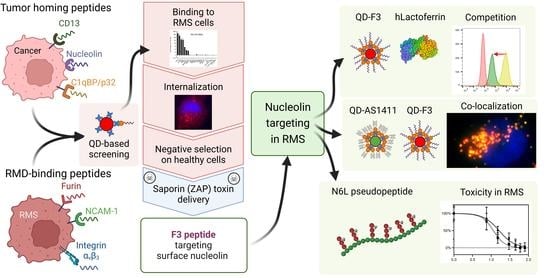
Quantum Dot-Based Screening Identifies F3 Peptide and Reveals Cell Surface Nucleolin as a Therapeutic Target for Rhabdomyosarcoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Lines
2.3. Preparation and Quality Controls of Quantum Dots Streptavidin-Biotin Conjugates
2.4. QD-Ligands Incubation with Cells
2.5. Fluorescence-Activated Cell Sorting (FACS)
2.6. Fluorescence Microscopy
2.7. ZAP-Conjugates’ Preparation and Cell Viability Assay
2.8. Lactoferrin Competition
2.9. Treatment with Pseudopeptide N6L
2.10. MTT Cell Viability Assay
3. Results
3.1. Validation of QD-Peptides Streptavidin-Biotin Conjugates
3.2. Screening of Peptide-Ligands Using QD Fluorescent Probe
3.3. Peptide-Mediated Internalization and Cytoplasmic Payload Delivery
3.4. Validation of Nucleolin as a Specific Target for RMS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ognjanovic, S.; Linabery, A.M.; Charbonneau, B.; Ross, J.A. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975–2005. Cancer 2009, 115, 4218–4226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loeb, D.M.; Thornton, K.; Shokek, O. Pediatric soft tissue sarcomas. Surg. Clin. N. Am. 2008, 88, 615–627. [Google Scholar] [CrossRef] [Green Version]
- Barr, F.G.; Smith, L.M.; Lynch, J.C.; Strzelecki, D.; Parham, D.M.; Qualman, S.J.; Breitfeld, P.P. Examination of gene fusion status in archival samples of alveolar rhabdomyosarcoma entered on the Intergroup Rhabdomyosarcoma Study-III trial: A report from the Children’s Oncology Group. J. Mol. Diagn. 2006, 8, 202–208. [Google Scholar] [CrossRef] [Green Version]
- Malempati, S.; Weigel, B.J.; Chi, Y.Y.; Tian, J.; Anderson, J.R.; Parham, D.M.; Teot, L.A.; Rodeberg, D.A.; Yock, T.I.; Shulkin, B.L.; et al. The addition of cixutumumab or temozolomide to intensive multiagent chemotherapy is feasible but does not improve outcome for patients with metastatic rhabdomyosarcoma: A report from the Children’s Oncology Group. Cancer 2019, 125, 290–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shern, J.F.; Selfe, J.; Izquierdo, E.; Patidar, R.; Chou, H.C.; Song, Y.K.; Yohe, M.E.; Sindiri, S.; Wei, J.; Wen, X.; et al. Genomic Classification and Clinical Outcome in Rhabdomyosarcoma: A Report From an International Consortium. J. Clin. Oncol. 2021, 39, 2859–2871. [Google Scholar] [CrossRef] [PubMed]
- Punyko, J.A.; Mertens, A.C.; Gurney, J.G.; Yasui, Y.; Donaldson, S.S.; Rodeberg, D.A.; Raney, R.B.; Stovall, M.; Sklar, C.A.; Robison, L.L.; et al. Long-term medical effects of childhood and adolescent rhabdomyosarcoma: A report from the childhood cancer survivor study. Pediatr. Blood Cancer 2005, 44, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Owosho, A.A.; Brady, P.; Wolden, S.L.; Wexler, L.H.; Antonescu, C.R.; Huryn, J.M.; Estilo, C.L. Long-term effect of chemotherapy-intensity-modulated radiation therapy (chemo-IMRT) on dentofacial development in head and neck rhabdomyosarcoma patients. Pediatr. Hematol. Oncol. 2016, 33, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Matsumura, Y.; Maeda, H. A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986, 46, 6387–6392. [Google Scholar]
- Hashida, M. Advocation and advancements of EPR effect theory in drug delivery science: A commentary. J. Control Release 2022, 346, 355–357. [Google Scholar] [CrossRef]
- Park, K. Questions on the role of the EPR effect in tumor targeting. J. Control Release 2013, 172, 391. [Google Scholar] [CrossRef]
- Sun, D.; Zhou, S.; Gao, W. What Went Wrong with Anticancer Nanomedicine Design and How to Make It Right. ACS Nano 2020, 14, 12281–12290. [Google Scholar] [CrossRef] [PubMed]
- Nag, O.K.; Delehanty, J.B. Active Cellular and Subcellular Targeting of Nanoparticles for Drug Delivery. Pharmaceutics 2019, 11, 543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araste, F.; Abnous, K.; Hashemi, M.; Taghdisi, S.M.; Ramezani, M.; Alibolandi, M. Peptide-based targeted therapeutics: Focus on cancer treatment. J. Control Release 2018, 292, 141–162. [Google Scholar] [CrossRef]
- Roveri, M.; Bernasconi, M.; Leroux, J.C.; Luciani, P. Peptides for tumor-specific drug targeting: State of the art and beyond. J. Mater. Chem. B 2017, 5, 4348–4364. [Google Scholar] [CrossRef] [PubMed]
- Roveri, M.; Pfohl, A.; Jaaks, P.; Alijaj, N.; Leroux, J.C.; Luciani, P.; Bernasconi, M. Prolonged circulation and increased tumor accumulation of liposomal vincristine in a mouse model of rhabdomyosarcoma. Nanomedicine 2017, 12, 1135–1151. [Google Scholar] [CrossRef]
- Hajdin, K.; D’Alessandro, V.; Niggli, F.K.; Schafer, B.W.; Bernasconi, M. Furin targeted drug delivery for treatment of rhabdomyosarcoma in a mouse model. PLoS ONE 2010, 5, e10445. [Google Scholar] [CrossRef] [Green Version]
- Bates, P.J.; Kahlon, J.B.; Thomas, S.D.; Trent, J.O.; Miller, D.M. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 1999, 274, 26369–26377. [Google Scholar] [CrossRef] [Green Version]
- Ko, M.H.; Kim, S.; Kang, W.J.; Lee, J.H.; Kang, H.; Moon, S.H.; Hwang, D.W.; Ko, H.Y.; Lee, D.S. In vitro derby imaging of cancer biomarkers using quantum dots. Small 2009, 5, 1207–1212. [Google Scholar] [CrossRef]
- Mittal, R.; Bruchez, M.P. Biotin-4-fluorescein based fluorescence quenching assay for determination of biotin binding capacity of streptavidin conjugated quantum dots. Bioconjug Chem. 2011, 22, 362–368. [Google Scholar] [CrossRef]
- Destouches, D.; Page, N.; Hamma-Kourbali, Y.; Machi, V.; Chaloin, O.; Frechault, S.; Birmpas, C.; Katsoris, P.; Beyrath, J.; Albanese, P.; et al. A simple approach to cancer therapy afforded by multivalent pseudopeptides that target cell-surface nucleoproteins. Cancer Res. 2011, 71, 3296–3305. [Google Scholar] [CrossRef] [Green Version]
- Sheikh, A.; Alhakamy, N.A.; Md, S.; Kesharwani, P. Recent Progress of RGD Modified Liposomes as Multistage Rocket Against Cancer. Front. Pharmacol. 2021, 12, 803304. [Google Scholar] [CrossRef]
- Ludwig, B.S.; Kessler, H.; Kossatz, S.; Reuning, U. RGD-Binding Integrins Revisited: How Recently Discovered Functions and Novel Synthetic Ligands (Re-)Shape an Ever-Evolving Field. Cancers 2021, 13, 1711. [Google Scholar] [CrossRef] [PubMed]
- Scherzinger-Laude, K.; Schonherr, C.; Lewrick, F.; Suss, R.; Francese, G.; Rossler, J. Treatment of neuroblastoma and rhabdomyosarcoma using RGD-modified liposomal formulations of patupilone (EPO906). Int. J. Nanomed. 2013, 8, 2197–2211. [Google Scholar] [CrossRef] [Green Version]
- Rengaswamy, V.; Zimmer, D.; Suss, R.; Rossler, J. RGD liposome-protamine-siRNA (LPR) nanoparticles targeting PAX3-FOXO1 for alveolar rhabdomyosarcoma therapy. J. Control Release 2016, 235, 319–327. [Google Scholar] [CrossRef]
- Zhou, M.; Ghosh, I. Quantum dots and peptides: A bright future together. Biopolymers 2007, 88, 325–339. [Google Scholar] [CrossRef]
- Christian, S.; Pilch, J.; Akerman, M.E.; Porkka, K.; Laakkonen, P.M.; Ruoslahti, E. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J. Cell Biol. 2003, 163, 871–878. [Google Scholar] [CrossRef] [Green Version]
- Porkka, K.; Laakkonen, P.; Hoffman, J.A.; Bernasconi, M.; Ruoslahti, E. A fragment of the HMGN2 protein homes to the nuclei of tumor cells and tumor endothelial cells in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 7444–7449. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, N.A.; Gomes-da-Silva, L.C.; Moura, V.; Simoes, S.; Moreira, J.N. Simultaneous active intracellular delivery of doxorubicin and C6-ceramide shifts the additive/antagonistic drug interaction of non-encapsulated combination. J. Control Release 2014, 196, 122–131. [Google Scholar] [CrossRef]
- Ellerby, H.M.; Arap, W.; Ellerby, L.M.; Kain, R.; Andrusiak, R.; Rio, G.D.; Krajewski, S.; Lombardo, C.R.; Rao, R.; Ruoslahti, E.; et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat. Med. 1999, 5, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Curnis, F.; Cattaneo, A.; Longhi, R.; Sacchi, A.; Gasparri, A.M.; Pastorino, F.; Di Matteo, P.; Traversari, C.; Bachi, A.; Ponzoni, M.; et al. Critical role of flanking residues in NGR-to-isoDGR transition and CD13/integrin receptor switching. J. Biol. Chem. 2010, 285, 9114–9123. [Google Scholar] [CrossRef] [Green Version]
- Laakkonen, P.; Porkka, K.; Hoffman, J.A.; Ruoslahti, E. A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nat. Med. 2002, 8, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Alberici, L.; Roth, L.; Sugahara, K.N.; Agemy, L.; Kotamraju, V.R.; Teesalu, T.; Bordignon, C.; Traversari, C.; Rizzardi, G.P.; Ruoslahti, E. De novo design of a tumor-penetrating peptide. Cancer Res. 2013, 73, 804–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, L.; Agemy, L.; Kotamraju, V.R.; Braun, G.; Teesalu, T.; Sugahara, K.N.; Hamzah, J.; Ruoslahti, E. Transtumoral targeting enabled by a novel neuropilin-binding peptide. Oncogene 2012, 31, 3754–3763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Girard, O.M.; Hanahan, D.; Mattrey, R.F.; Ruoslahti, E. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 2009, 16, 510–520. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Feng, M.; Hu, J.; Chen, C.; Wang, J.; Wang, X.; Xu, H.; Lu, J.R. Designed short RGD peptides for one-pot aqueous synthesis of integrin-binding CdTe and CdZnTe quantum dots. ACS Appl. Mater. Interfaces 2012, 4, 6362–6370. [Google Scholar] [CrossRef]
- Haubner, R.; Gratias, R.; Diefenbach, B.; Goodman, S.L.; Jonczyk, A.; Kessler, H. Structural and functional aspects of RGD-containing cyclic pentapeptides as highly potent and selective integrin αvβ3 antagonists. J. Am. Chem. Soc. 1996, 118, 7461–7472. [Google Scholar] [CrossRef]
- Chen, X.; Park, R.; Shahinian, A.H.; Tohme, M.; Khankaldyyan, V.; Bozorgzadeh, M.H.; Bading, J.R.; Moats, R.; Laug, W.E.; Conti, P.S. 18F-labeled RGD peptide: Initial evaluation for imaging brain tumor angiogenesis. Nucl. Med. Biol. 2004, 31, 179–189. [Google Scholar] [CrossRef]
- Van Hagen, P.M.; Breeman, W.A.; Bernard, H.F.; Schaar, M.; Mooij, C.M.; Srinivasan, A.; Schmidt, M.A.; Krenning, E.P.; de Jong, M. Evaluation of a radiolabelled cyclic DTPA-RGD analogue for tumour imaging and radionuclide therapy. Int. J. Cancer 2000, 90, 186–198. [Google Scholar] [CrossRef]
- Witt, H.; Hajdin, K.; Iljin, K.; Greiner, O.; Niggli, F.K.; Schafer, B.W.; Bernasconi, M. Identification of a rhabdomyosarcoma targeting peptide by phage display with sequence similarities to the tumour lymphatic-homing peptide LyP-1. Int. J. Cancer 2009, 124, 2026–2032. [Google Scholar] [CrossRef]
- Kiryushko, D.; Kofoed, T.; Skladchikova, G.; Holm, A.; Berezin, V.; Bock, E. A synthetic peptide ligand of neural cell adhesion molecule (NCAM), C3d, promotes neuritogenesis and synaptogenesis and modulates presynaptic function in primary cultures of rat hippocampal neurons. J. Biol. Chem. 2003, 278, 12325–12334. [Google Scholar] [CrossRef] [Green Version]
- Ronn, L.C.; Olsen, M.; Ostergaard, S.; Kiselyov, V.; Berezin, V.; Mortensen, M.T.; Lerche, M.H.; Jensen, P.H.; Soroka, V.; Saffell, J.L.; et al. Identification of a neuritogenic ligand of the neural cell adhesion molecule using a combinatorial library of synthetic peptides. Nat. Biotechnol. 1999, 17, 1000–1005. [Google Scholar] [CrossRef] [PubMed]
- Markovsky, E.; Vax, E.; Ben-Shushan, D.; Eldar-Boock, A.; Shukrun, R.; Yeini, E.; Barshack, I.; Caspi, R.; Harari-Steinberg, O.; Pode-Shakked, N.; et al. Wilms tumor NCAM-expressing cancer stem cells as potential therapeutic target for polymeric nanomedicine. Mol. Cancer Ther. 2017, 16, 2462–2472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heimann, A.S.; Gomes, I.; Dale, C.S.; Pagano, R.L.; Gupta, A.; de Souza, L.L.; Luchessi, A.D.; Castro, L.M.; Giorgi, R.; Rioli, V.; et al. Hemopressin is an inverse agonist of CB1 cannabinoid receptors. Proc. Natl. Acad. Sci. USA 2007, 104, 20588–20593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Zhao, R.; Wu, X.; Sun, Y.; Yao, M.; Li, J.; Xu, Y.; Gu, J. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J. 2005, 19, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Engler, J.A.; Collawn, J.F.; Moore, B.A. Receptor mediated uptake of peptides that bind the human transferrin receptor. Eur. J. Biochem. 2001, 268, 2004–2012. [Google Scholar] [CrossRef]
- Lu, Y.; Jiang, W.; Wu, X.; Huang, S.; Huang, Z.; Shi, Y.; Dai, Q.; Chen, J.; Ren, F.; Gao, S. Peptide T7-modified polypeptide with disulfide bonds for targeted delivery of plasmid DNA for gene therapy of prostate cancer. Int. J. Nanomed. 2018, 13, 6913–6927. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Wang, Q.; Yu, Q.; Qiu, Y.; Mei, L.; Wan, D.; Wang, X.; Li, M.; He, Q. A stabilized retro-inverso peptide ligand of transferrin receptor for enhanced liposome-based hepatocellular carcinoma-targeted drug delivery. Acta Biomater. 2019, 83, 379–389. [Google Scholar] [CrossRef]
- Devulapally, R.; Sekar, N.M.; Sekar, T.V.; Foygel, K.; Massoud, T.F.; Willmann, J.K.; Paulmurugan, R. Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy. ACS Nano 2015, 9, 2290–2302. [Google Scholar] [CrossRef] [Green Version]
- Stirpe, F.; Gasperi-Campani, A.; Barbieri, L.; Falasca, A.; Abbondanza, A.; Stevens, W.A. Ribosome-inactivating proteins from the seeds of Saponaria officinalis L. (soapwort), of Agrostemma githago L. (corn cockle) and of Asparagus officinalis L. (asparagus), and from the latex of Hura crepitans L. (sandbox tree). Biochem. J. 1983, 216, 617–625. [Google Scholar] [CrossRef] [Green Version]
- Ancheta, L.R.; Shramm, P.A.; Bouajram, R.; Higgins, D.; Lappi, D.A. Saporin as a commercial reagent: Its uses and unexpected impacts in the biological sciences-tools from the plant kingdom. Toxins 2022, 14, 184. [Google Scholar] [CrossRef]
- Huang, Y.; Shi, H.; Zhou, H.; Song, X.; Yuan, S.; Luo, Y. The angiogenic function of nucleolin is mediated by vascular endothelial growth factor and nonmuscle myosin. Blood 2006, 107, 3564–3571. [Google Scholar] [CrossRef]
- Watanabe, T.; Hirano, K.; Takahashi, A.; Yamaguchi, K.; Beppu, M.; Fujiki, H.; Suganuma, M. Nucleolin on the cell surface as a new molecular target for gastric cancer treatment. Biol. Pharm. Bull. 2010, 33, 796–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Song, N.; Liu, C.; He, T.; Zhuo, W.; He, X.; Chen, Y.; Song, X.; Fu, Y.; Luo, Y. Heat shock cognate 70 regulates the translocation and angiogenic function of nucleolin. Arter. Thromb. Vasc. Biol. 2012, 32, e126–e134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.C.; Hu, T.H.; Huang, C.C.; Kung, M.L.; Chu, T.H.; Yi, L.N.; Huang, S.T.; Chan, H.H.; Chuang, J.H.; Liu, L.F.; et al. Hepatoma-derived growth factor/nucleolin axis as a novel oncogenic pathway in liver carcinogenesis. Oncotarget 2015, 6, 16253–16270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krust, B.; El Khoury, D.; Nondier, I.; Soundaramourty, C.; Hovanessian, A.G. Targeting surface nucleolin with multivalent HB-19 and related Nucant pseudopeptides results in distinct inhibitory mechanisms depending on the malignant tumor cell type. BMC Cancer 2011, 11, 333. [Google Scholar] [CrossRef] [Green Version]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef] [Green Version]
- Destouches, D.; El Khoury, D.; Hamma-Kourbali, Y.; Krust, B.; Albanese, P.; Katsoris, P.; Guichard, G.; Briand, J.P.; Courty, J.; Hovanessian, A.G. Suppression of tumor growth and angiogenesis by a specific antagonist of the cell-surface expressed nucleolin. PLoS ONE 2008, 3, e2518. [Google Scholar] [CrossRef] [Green Version]
- Callebaut, C.; Blanco, J.; Benkirane, N.; Krust, B.; Jacotot, E.; Guichard, G.; Seddiki, N.; Svab, J.; Dam, E.; Muller, S.; et al. Identification of V3 loop-binding proteins as potential receptors implicated in the binding of HIV particles to CD4(+) cells. J. Biol. Chem. 1998, 273, 21988–21997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girvan, A.C.; Teng, Y.; Casson, L.K.; Thomas, S.D.; Juliger, S.; Ball, M.W.; Klein, J.B.; Pierce, W.M., Jr.; Barve, S.S.; Bates, P.J. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006, 5, 1790–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Reyes, E.M.; Teng, Y.; Bates, P.J. A new paradigm for aptamer therapeutic AS1411 action: Uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010, 70, 8617–8629. [Google Scholar] [CrossRef]
- Xing, H.; Tang, L.; Yang, X.; Hwang, K.; Wang, W.; Yin, Q.; Wong, N.Y.; Dobrucki, L.W.; Yasui, N.; Katzenellenbogen, J.A.; et al. Selective Delivery of an Anticancer Drug with Aptamer-Functionalized Liposomes to Breast Cancer Cells in Vitro and in Vivo. J. Mater. Chem. B 2013, 1, 5288–5297. [Google Scholar] [CrossRef]
- Li, S.; Jiang, Q.; Liu, S.; Zhang, Y.; Tian, Y.; Song, C.; Wang, J.; Zou, Y.; Anderson, G.J.; Han, J.Y.; et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018, 36, 258–264. [Google Scholar] [CrossRef]
- Legrand, D.; Vigie, K.; Said, E.A.; Elass, E.; Masson, M.; Slomianny, M.C.; Carpentier, M.; Briand, J.P.; Mazurier, J.; Hovanessian, A.G. Surface nucleolin participates in both the binding and endocytosis of lactoferrin in target cells. Eur. J. Biochem. 2004, 271, 303–317. [Google Scholar] [CrossRef]
- Alibolandi, M.; Abnous, K.; Ramezani, M.; Hosseinkhani, H.; Hadizadeh, F. Synthesis of AS1411-aptamer-conjugated CdTe quantum dots with high fluorescence strength for probe labeling tumor cells. J. Fluoresc. 2014, 24, 1519–1529. [Google Scholar] [CrossRef]
- Zheng, S.; Zhang, M.; Bai, H.; He, M.; Dong, L.; Cai, L.; Zhao, M.; Wang, Q.; Xu, K.; Li, J. Preparation of AS1411 Aptamer Modified Mn-MoS2 QDs for Targeted MR Imaging and Fluorescence Labelling of Renal Cell Carcinoma. Int. J. Nanomed. 2019, 14, 9513–9524. [Google Scholar] [CrossRef] [Green Version]
- Take, M.; Tsutsui, J.; Obama, H.; Ozawa, M.; Nakayama, T.; Maruyama, I.; Arima, T.; Muramatsu, T. Identification of nucleolin as a binding protein for midkine (MK) and heparin-binding growth associated molecule (HB-GAM). J. Biochem. 1994, 116, 1063–1068. [Google Scholar] [CrossRef]
- Bulin, A.L.; Hasan, T. Spatiotemporal Tracking of Different Cell Populations in Cancer Organoid Models for Investigations on Photodynamic Therapy. Methods Mol. Biol. 2022, 2451, 81–90. [Google Scholar] [CrossRef]
- Vinduska, V.; Gallops, C.E.; O’Connor, R.; Wang, Y.; Huang, X. Exosomal Surface Protein Detection with Quantum Dots and Immunomagnetic Capture for Cancer Detection. Nanomaterials 2021, 11, 1853. [Google Scholar] [CrossRef]
- Kuo, C.W.; Chueh, D.Y.; Chen, P. Real-time in vivo imaging of subpopulations of circulating tumor cells using antibody conjugated quantum dots. J. Nanobiotech. 2019, 17, 26. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.Y.; Cheng, B.R.; He, Z.B.; Wang, S.Y.; Wang, Z.M.; Sun, M.; Song, H.B.; Fang, Y.; Chen, F.F.; Xiong, B. Capture and Identification of Heterogeneous Circulating Tumor Cells Using Transparent Nanomaterials and Quantum Dots-Based Multiplexed Imaging. J. Cancer 2016, 7, 69–79. [Google Scholar] [CrossRef]
- Min, H.; Jo, S.M.; Kim, H.S. Efficient capture and simple quantification of circulating tumor cells using quantum dots and magnetic beads. Small 2015, 11, 2536–2542. [Google Scholar] [CrossRef]
- Lam, P.Y.; Hillyar, C.R.; Able, S.; Vallis, K.A. Synthesis and evaluation of an (18) F-labeled derivative of F3 for targeting surface-expressed nucleolin in cancer and tumor endothelial cells. J. Label. Compd. Radiopharm. 2016, 59, 492–499. [Google Scholar] [CrossRef] [Green Version]
- Palmieri, D.; Richmond, T.; Piovan, C.; Sheetz, T.; Zanesi, N.; Troise, F.; James, C.; Wernicke, D.; Nyei, F.; Gordon, T.J.; et al. Human anti-nucleolin recombinant immunoagent for cancer therapy. Proc. Natl. Acad. Sci. USA 2015, 112, 9418–9423. [Google Scholar] [CrossRef] [Green Version]
- Diamantopoulou, Z.; Gilles, M.E.; Sader, M.; Cossutta, M.; Vallee, B.; Houppe, C.; Habert, D.; Brissault, B.; Leroy, E.; Maione, F.; et al. Multivalent cationic pseudopeptide polyplexes as a tool for cancer therapy. Oncotarget 2017, 8, 90108–90122. [Google Scholar] [CrossRef] [Green Version]
- Jia, W.; Yao, Z.; Zhao, J.; Guan, Q.; Gao, L. New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sci. 2017, 186, 1–10. [Google Scholar] [CrossRef]
- Ginisty, H.; Sicard, H.; Roger, B.; Bouvet, P. Structure and functions of nucleolin. J. Cell Sci. 1999, 112 Pt 6, 761–772. [Google Scholar] [CrossRef]
- Ugrinova, I.; Petrova, M.; Chalabi-Dchar, M.; Bouvet, P. Multifaceted Nucleolin Protein and Its Molecular Partners in Oncogenesis. Adv. Protein Chem. Struct. Biol. 2018, 111, 133–164. [Google Scholar] [CrossRef]
- Koutsioumpa, M.; Papadimitriou, E. Cell surface nucleolin as a target for anti-cancer therapies. Recent Pat. Anti-Cancer Drug Discov. 2014, 9, 137–152. [Google Scholar] [CrossRef]
- Romano, S.; Fonseca, N.; Simoes, S.; Goncalves, J.; Moreira, J.N. Nucleolin-based targeting strategies for cancer therapy: From targeted drug delivery to cytotoxic ligands. Drug Discov. Today 2019, 24, 1985–2001. [Google Scholar] [CrossRef]
- Carvalho, L.S.; Goncalves, N.; Fonseca, N.A.; Moreira, J.N. Cancer Stem Cells and Nucleolin as Drivers of Carcinogenesis. Pharmaceuticals 2021, 14, 60. [Google Scholar] [CrossRef]
- Ferrara, B.; Belbekhouche, S.; Habert, D.; Houppe, C.; Vallee, B.; Bourgoin-Voillard, S.; Cohen, J.L.; Cascone, I.; Courty, J. Cell surface nucleolin as active bait for nanomedicine in cancer therapy: A promising option. Nanotechnology 2021, 32, 322001. [Google Scholar] [CrossRef]
- Tonello, F.; Massimino, M.L.; Peggion, C. Nucleolin: A cell portal for viruses, bacteria, and toxins. Cell Mol. Life Sci. 2022, 79, 271. [Google Scholar] [CrossRef]
- Karamchand, L.; Kim, G.; Wang, S.; Hah, H.J.; Ray, A.; Jiddou, R.; Koo Lee, Y.E.; Philbert, M.A.; Kopelman, R. Modulation of hydrogel nanoparticle intracellular trafficking by multivalent surface engineering with tumor targeting peptide. Nanoscale 2013, 5, 10327–10344. [Google Scholar] [CrossRef] [Green Version]
- Su, P.Y.; Wang, Y.F.; Huang, S.W.; Lo, Y.C.; Wang, Y.H.; Wu, S.R.; Shieh, D.B.; Chen, S.H.; Wang, J.R.; Lai, M.D.; et al. Cell surface nucleolin facilitates enterovirus 71 binding and infection. J. Virol. 2015, 89, 4527–4538. [Google Scholar] [CrossRef] [Green Version]
- Hovanessian, A.G.; Puvion-Dutilleul, F.; Nisole, S.; Svab, J.; Perret, E.; Deng, J.S.; Krust, B. The cell-surface-expressed nucleolin is associated with the actin cytoskeleton. Exp. Cell Res. 2000, 261, 312–328. [Google Scholar] [CrossRef]
- Nohira, N.; Shinji, S.; Nakamura, S.; Nihashi, Y.; Shimosato, T.; Takaya, T. Myogenetic oligodeoxynucleotides as anti-nucleolin aptamers inhibit the growth of embryonal rhabdomyosarcoma cells. bioRxiv 2021. [Google Scholar] [CrossRef]
- Willmer, T.; Damerell, V.; Smyly, S.; Sims, D.; Du Toit, M.; Ncube, S.; Sinkala, M.; Govender, D.; Sturrock, E.; Blackburn, J.M.; et al. Targeting the oncogenic TBX3:nucleolin complex to treat multiple sarcoma subtypes. Am. J. Cancer Res. 2021, 11, 5680–5700. [Google Scholar]
- Sims, D.; Maranyane, H.M.; Damerell, V.; Govender, D.; Isaacs, A.W.; Peres, J.; Prince, S. The c-Myc/AKT1/TBX3 Axis Is Important to Target in the Treatment of Embryonal Rhabdomyosarcoma. Cancers 2020, 12, 501. [Google Scholar] [CrossRef] [Green Version]
- Rammal, G.; Fahs, A.; Kobeissy, F.; Mechref, Y.; Zhao, J.; Zhu, R.; Diab-Assaf, M.; Saab, R.; Ghayad, S.E. Proteomic Profiling of Rhabdomyosarcoma-Derived Exosomes Yield Insights into Their Functional Role in Paracrine Signaling. J. Proteome Res. 2019, 18, 3567–3579. [Google Scholar] [CrossRef]
- Benedetti, E.; Antonosante, A.; d’Angelo, M.; Cristiano, L.; Galzio, R.; Destouches, D.; Florio, T.M.; Dhez, A.C.; Astarita, C.; Cinque, B.; et al. Nucleolin antagonist triggers autophagic cell death in human glioblastoma primary cells and decreased in vivo tumor growth in orthotopic brain tumor model. Oncotarget 2015, 6, 42091–42104. [Google Scholar] [CrossRef] [Green Version]
- Gilles, M.E.; Maione, F.; Cossutta, M.; Carpentier, G.; Caruana, L.; Di Maria, S.; Houppe, C.; Destouches, D.; Shchors, K.; Prochasson, C.; et al. Nucleolin Targeting Impairs the Progression of Pancreatic Cancer and Promotes the Normalization of Tumor Vasculature. Cancer Res. 2016, 76, 7181–7193. [Google Scholar] [CrossRef]
- Fonseca, N.A.; Gregório, A.C.; Mendes, V.M.; Lopes, R.; Abreu, T.; Gonçalves, N.; Manadas, B.; Lacerda, M.; Figueiredo, P.; Pereira, M.; et al. GMP-grade nanoparticle targeted to nucleolin downregulates tumor molecular signature, blocking growth and invasion, at low systemic exposure. Nano Today 2021, 37, 101095. [Google Scholar] [CrossRef]
- Brignole, C.; Bensa, V.; Fonseca, N.A.; Del Zotto, G.; Bruno, S.; Cruz, A.F.; Malaguti, F.; Carlini, B.; Morandi, F.; Calarco, E.; et al. Cell surface Nucleolin represents a novel cellular target for neuroblastoma therapy. J. Exp. Clin. Cancer Res. 2021, 40, 180. [Google Scholar] [CrossRef] [PubMed]
- Jaaks, P.; D’Alessandro, V.; Grob, N.; Buel, S.; Hajdin, K.; Schafer, B.W.; Bernasconi, M. The Proprotein Convertase Furin Contributes to Rhabdomyosarcoma Malignancy by Promoting Vascularization, Migration and Invasion. PLoS ONE 2016, 11, e0161396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaaks, P.; Meier, G.; Alijaj, N.; Brack, E.; Bode, P.; Koscielniak, E.; Wachtel, M.; Schafer, B.W.; Bernasconi, M. The proprotein convertase furin is required to maintain viability of alveolar rhabdomyosarcoma cells. Oncotarget 2016, 7, 76743–76755. [Google Scholar] [CrossRef] [Green Version]
- Phimister, E.G.; Culverwell, A.; Patel, K.; Kemshead, J.T. Tissue-specific expression of neural cell adhesion molecule (NCAM) may allow differential diagnosis of neuroblastoma from embryonal rhabdomyosarcoma. Eur. J. Cancer 1994, 30A, 1552–1558. [Google Scholar] [CrossRef]
- Oesch, S.; Walter, D.; Wachtel, M.; Pretre, K.; Salazar, M.; Guzman, M.; Velasco, G.; Schafer, B.W. Cannabinoid receptor 1 is a potential drug target for treatment of translocation-positive rhabdomyosarcoma. Mol. Cancer Ther. 2009, 8, 1838–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Giovanni, C.; Landuzzi, L.; Palladini, A.; Nicoletti, G.; Nanni, P.; Lollini, P.L. HER Tyrosine Kinase Family and Rhabdomyosarcoma: Role in Onset and Targeted Therapy. Cells 2021, 10, 1808. [Google Scholar] [CrossRef]
- Ganti, R.; Skapek, S.X.; Zhang, J.; Fuller, C.E.; Wu, J.; Billups, C.A.; Breitfeld, P.P.; Dalton, J.D.; Meyer, W.H.; Khoury, J.D. Expression and genomic status of EGFR and ErbB-2 in alveolar and embryonal rhabdomyosarcoma. Mod. Pathol. 2006, 19, 1213–1220. [Google Scholar] [CrossRef] [Green Version]
- Falvo, E.; Damiani, V.; Conti, G.; Boschi, F.; Messana, K.; Giacomini, P.; Milella, M.; De Laurenzi, V.; Morea, V.; Sala, G.; et al. High activity and low toxicity of a novel CD71-targeting nanotherapeutic named The-0504 on preclinical models of several human aggressive tumors. J. Exp. Clin. Cancer Res. 2021, 40, 63. [Google Scholar] [CrossRef] [PubMed]
- Oh, F.; Todhunter, D.; Taras, E.; Vallera, D.A.; Borgatti, A. Targeting EGFR and uPAR on human rhabdomyosarcoma, osteosarcoma, and ovarian adenocarcinoma with a bispecific ligand-directed toxin. Clin. Pharmacol. 2018, 10, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Pilbeam, K.; Wang, H.; Taras, E.; Bergerson, R.J.; Ettestad, B.; DeFor, T.; Borgatti, A.; Vallera, D.A.; Verneris, M.R. Targeting pediatric sarcoma with a bispecific ligand immunotoxin targeting urokinase and epidermal growth factor receptors. Oncotarget 2018, 9, 11938–11947. [Google Scholar] [CrossRef] [PubMed]
- Kessler, T.; Baumeier, A.; Brand, C.; Grau, M.; Angenendt, L.; Harrach, S.; Stalmann, U.; Schmidt, L.H.; Gosheger, G.; Hardes, J.; et al. Aminopeptidase N (CD13): Expression, Prognostic Impact, and Use as Therapeutic Target for Tissue Factor Induced Tumor Vascular Infarction in Soft Tissue Sarcoma. Transl. Oncol. 2018, 11, 1271–1282. [Google Scholar] [CrossRef]
- Brohl, A.S.; Sindiri, S.; Wei, J.S.; Milewski, D.; Chou, H.C.; Song, Y.K.; Wen, X.; Kumar, J.; Reardon, H.V.; Mudunuri, U.S.; et al. Immuno-transcriptomic profiling of extracranial pediatric solid malignancies. Cell Rep. 2021, 37, 110047. [Google Scholar] [CrossRef]
- Hayes, M.N.; McCarthy, K.; Jin, A.; Oliveira, M.L.; Iyer, S.; Garcia, S.P.; Sindiri, S.; Gryder, B.; Motala, Z.; Nielsen, G.P.; et al. Vangl2/RhoA Signaling Pathway Regulates Stem Cell Self-Renewal Programs and Growth in Rhabdomyosarcoma. Cell Stem. Cell 2018, 22, 414–427.e416. [Google Scholar] [CrossRef]





| Targets Expressed on Tumor Blood and/or Lymphatic Vessels as Well as on Tumors | |||
|---|---|---|---|
| Target | Peptide | Sequence | Ref. |
| Nucleolin | F3 (29 aa) | EPQRRSARLSAKPAPPKPEPKPKKAPAKK | [25,26,27] |
| CD13 | NGR-5 | CNGRC * | [28] |
| NGR-18 | CNGRCGVRSSSRTPSDKY | [29] | |
| p32 | Lyp-1 | CGNKRTRGC | [30] |
| CD13, Neuropilin-1 | iNGR | CRNGRGPDC | [31] |
| p32, Neuropilin-1, | tLyp-1 | CGNKRTR | [32] |
| Integrin αvβ3, Neuropilin-1 | iRGD | CRGDKGPDC | [33] |
| Targets Expressed on Rhabdomyosarcoma | |||
| Target | Peptide | Sequence | Ref. |
| Integrin αvβ3 | Linear RGD | CRGDS | [34] |
| Cyclic RGD | cRGDyK | [35,36,37] | |
| RMS-I | CQQSNRGDRKRC * | [38] | |
| Furin | TmR | KRDRGGGCMGTINTRTRRC * | [15] |
| shTmR | KRDRCMGTINTRTRRC * | This work | |
| RMS-P3-3G | GGGCMGTINTRTRRC * | [16] | |
| CmR (neg. ctr.) | KRDRGGGCMGTINTATAAC * | [15] | |
| Neural Cell Adhesion Molecule 1 (NCAM-1) | NTP | ASKKPKRNIKA | [39,40,41] |
| Cannabinoid Receptor 1 (CB1) | Hemopressin | PVNFKFLSH | [42] |
| Epidermal Growth Factor Receptor (EGFR)/ErbB1 | GE11 | YHWYGYTPQNVI | [43] |
| Transferrin Receptor 1 (TFR1) | T7 | HAIYPRH | [44,45] |
| iso-d-T7 | d-(HRPYIAH) | [46] | |
| Urokinase Plasminogen Activator Receptor (uPAR) | uPA | VSNKYFSNIHWGC | [47] |
| Target undetermined | RMS-II | CMGNKRSAKRPC * | [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dzhumashev, D.; Timpanaro, A.; Ali, S.; De Micheli, A.J.; Mamchaoui, K.; Cascone, I.; Rössler, J.; Bernasconi, M. Quantum Dot-Based Screening Identifies F3 Peptide and Reveals Cell Surface Nucleolin as a Therapeutic Target for Rhabdomyosarcoma. Cancers 2022, 14, 5048. https://doi.org/10.3390/cancers14205048
Dzhumashev D, Timpanaro A, Ali S, De Micheli AJ, Mamchaoui K, Cascone I, Rössler J, Bernasconi M. Quantum Dot-Based Screening Identifies F3 Peptide and Reveals Cell Surface Nucleolin as a Therapeutic Target for Rhabdomyosarcoma. Cancers. 2022; 14(20):5048. https://doi.org/10.3390/cancers14205048
Chicago/Turabian StyleDzhumashev, Dzhangar, Andrea Timpanaro, Safa Ali, Andrea J. De Micheli, Kamel Mamchaoui, Ilaria Cascone, Jochen Rössler, and Michele Bernasconi. 2022. "Quantum Dot-Based Screening Identifies F3 Peptide and Reveals Cell Surface Nucleolin as a Therapeutic Target for Rhabdomyosarcoma" Cancers 14, no. 20: 5048. https://doi.org/10.3390/cancers14205048
APA StyleDzhumashev, D., Timpanaro, A., Ali, S., De Micheli, A. J., Mamchaoui, K., Cascone, I., Rössler, J., & Bernasconi, M. (2022). Quantum Dot-Based Screening Identifies F3 Peptide and Reveals Cell Surface Nucleolin as a Therapeutic Target for Rhabdomyosarcoma. Cancers, 14(20), 5048. https://doi.org/10.3390/cancers14205048