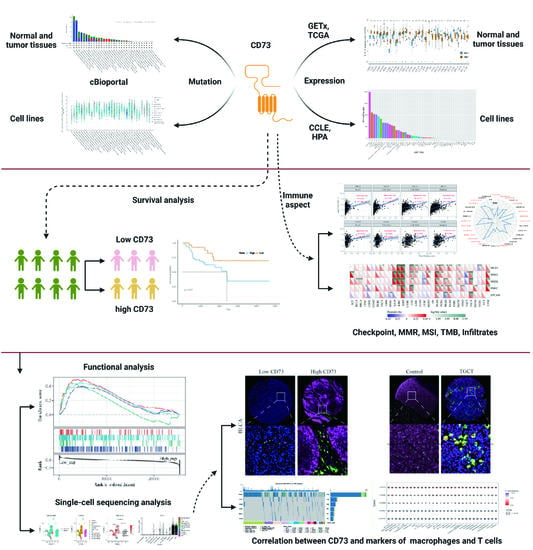
Identification of CD73 as a Novel Biomarker Encompassing the Tumor Microenvironment, Prognosis, and Therapeutic Responses in Various Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collecting and Organizing
2.2. Identification of CD73-Related Features
2.3. Single-Cell Sequencing Analysis
2.4. Multiple Fluorescent Staining
2.5. Statistical Analysis
3. Results
3.1. CD73 Expression in Tumor Tissues, Counterparts and Cell Lines
3.2. Mutational Aspects and Prognostic Role of CD73
3.3. Immune Characteristics of CD73 in the TME
3.4. CD73 Correlated with Checkpoints, MMR Markers, TMB, and MSI
3.5. Functional Analysis Based on CD73 Expression
3.6. Single Cell Sequencing to Reveal CD73 Expression on Tumor and Stromal Cells
3.7. Single Cell Sequencing to Analyze CD73 Expression and Related Signaling Pathways in GBM
3.8. Correlation between Macrophages, T Cells, and CD73 Expression
3.9. Potential Therapeutic Values of CD73 in Immunotherapy Cohorts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 1, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Sanchez-Salas, R.; Peske, J.D.; Vano, Y.; Becht, E.; Petitprez, F.; Validire, P.; Ingels, A.; Cathelineau, X.; Fridman, W.H.; et al. The clinical role of the TME in solid cancer. Br. J. Cancer 2019, 1, 45–53. [Google Scholar] [CrossRef]
- Labani-Motlagh, A.; Ashja-Mahdavi, M.; Loskog, A. The Tumor Microenvironment: A Milieu Hindering and Obstructing Antitumor Immune Responses. Front. Immunol. 2020, 1, 940. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 1, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Vinay, D.S.; Ryan, E.P.; Pawelec, G.; Talib, W.H.; Stagg, J.; Elkord, E.; Lichtor, T.; Decker, W.K.; Whelan, R.L.; Kumara, H.; et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 2015, 35, S185–S198. [Google Scholar] [CrossRef]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 1, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.; Xu, C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020, 1, 660–669. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 1, 1350–1355. [Google Scholar] [CrossRef] [Green Version]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 1, 711–723. [Google Scholar] [CrossRef]
- Schachter, J.; Ribas, A.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus ipilimumab for advanced melanoma: Final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 2017, 1, 1853–1862. [Google Scholar] [CrossRef]
- Zhang, H.; Dai, Z.; Wu, W.; Wang, Z.; Zhang, N.; Zhang, L.; Zeng, W.J.; Liu, Z.; Cheng, Q. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res. 2021, 1, 184. [Google Scholar] [CrossRef] [PubMed]
- Knapp, K.; Zebisch, M.; Pippel, J.; El-Tayeb, A.; Muller, C.E.; Strater, N. Crystal structure of the human ecto-5′-nucleotidase (CD73): Insights into the regulation of purinergic signaling. Structure 2012, 1, 2161–2173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Junger, W.G. Immune cell regulation by autocrine purinergic signalling. Nat. Rev. Immunol. 2011, 1, 201–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serra, S.; Horenstein, A.L.; Vaisitti, T.; Brusa, D.; Rossi, D.; Laurenti, L.; D’Arena, G.; Coscia, M.; Tripodo, C.; Inghirami, G.; et al. CD73-generated extracellular adenosine in chronic lymphocytic leukemia creates local conditions counteracting drug-induced cell death. Blood 2011, 1, 6141–6152. [Google Scholar] [CrossRef] [PubMed]
- Buisseret, L.; Pommey, S.; Allard, B.; Garaud, S.; Bergeron, M.; Cousineau, I.; Ameye, L.; Bareche, Y.; Paesmans, M.; Crown, J.P.A.; et al. Clinical significance of CD73 in triple-negative breast cancer: Multiplex analysis of a phase III clinical trial. Ann. Oncol. 2018, 1, 1056–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koivisto, M.K.; Tervahartiala, M.; Kenessey, I.; Jalkanen, S.; Bostrom, P.J.; Salmi, M. Cell-type-specific CD73 expression is an independent prognostic factor in bladder cancer. Carcinogenesis 2019, 1, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Walle, T.; Cornish, A.E.; Basu, S.; Anandhan, S.; Fernandez, I.; Vence, L.; Blando, J.; Zhao, H.; Yadav, S.S.; et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat. Med. 2020, 1, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.B.; Phan, L.H.; Villarreal, O.E.; Bowser, J.L. CD73’s Potential as an Immunotherapy Target in Gastrointestinal Cancers. Front. Immunol. 2020, 1, 508. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 1, 1113–1120. [Google Scholar]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 1, 1318–1330. [Google Scholar]
- Zhang, J.; Wang, Z.; Zhang, X.; Dai, Z.; Zhi-Peng, W.; Yu, J.; Peng, Y.; Wu, W.; Zhang, N.; Luo, P.; et al. Large-Scale Single-Cell and Bulk Sequencing Analyses Reveal the Prognostic Value and Immune Aspects of CD147 in Pan-Cancer. Front. Immunol. 2022, 1, 810471. [Google Scholar] [CrossRef] [PubMed]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R., 3rd; Kalocsay, M.; Jane-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 1, 387–402.e16. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 1, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 1, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goel, M.K.; Khanna, P.; Kishore, J. Understanding survival analysis: Kaplan-Meier estimate. Int. J. Ayurveda Res. 2010, 1, 274–278. [Google Scholar]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 1, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 1, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Horinaka, A.; Kim, Y.H.; Kimura, A.; Iwamoto, E.; Masaki, T.; Ichijo, T.; Sato, S. Changes in the predicted function of the rumen bacterial community of Japanese Black beef cattle during the fattening stages according to Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. J. Vet. Med. Sci. 2021, 1, 1098–1106. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.X.; Li, K.R.; Zhang, W.B.; Wan, C.X.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.X.; Wong, C.J.; Yang, L.; Ouardaoui, N.; Li, D.A.; Zhang, W.B.; Gu, S.Q.; Zhang, Y.; Liu, Y.; Wang, X.Q.; et al. TISMO: Syngeneic mouse tumor database to model tumor immunity and immunotherapy response. Nucleic Acids Res. 2022, 50, D1391–D1397. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Wang, Z.; Zhang, X.; Dai, Z.; Wu, W.; Zhang, N.; Liu, Z.; Zhang, J.; Luo, P.; et al. Identify the Prognostic and Immune Profile of VSIR in the Tumor Microenvironment: A Pan-Cancer Analysis. Front. Cell Dev. Biol. 2022, 1, 821649. [Google Scholar] [CrossRef]
- Chung, W.; Eum, H.H.; Lee, H.O.; Lee, K.M.; Lee, H.B.; Kim, K.T.; Ryu, H.S.; Kim, S.; Lee, J.E.; Park, Y.H.; et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017, 8, 15081. [Google Scholar] [CrossRef] [Green Version]
- Karaayvaz, M.; Cristea, S.; Gillespie, S.M.; Patel, A.P.; Mylvaganam, R.; Luo, C.C.; Specht, M.C.; Bernstein, B.E.; Michor, F.; Ellisen, L.W. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018, 9, 3588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, V.; Ramnarayanan, K.; Sundar, R.; Padmanabhan, N.; Srivastava, S.; Koiwa, M.; Yasuda, T.; Koh, V.; Huang, K.K.; Tay, S.T.; et al. Single-Cell Atlas of Lineage States, Tumor Microenvironment, and Subtype-Specific Expression Programs in Gastric Cancer. Cancer Discov. 2022, 1, 670–691. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 1, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; He, J.; Chen, R.; Wang, Z.; Dai, Z.; Liang, X.; Wu, W.; Luo, P.; Zhang, J.; Peng, Y.; et al. Pan-Cancer Analysis of the Immunological Role of PDIA5: A Potential Target for Immunotherapy. Front. Immunol. 2022, 1, 881722. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Novakovic, N.; Hua, Y.; Keep, R.F.; Xi, G. Role of lipocalin-2 in extracellular peroxiredoxin 2-induced brain swelling, inflammation and neuronal death. Exp. Neurol. 2021, 1, 113521. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, H.; Wang, Z.; Dai, Z.; Zhang, X.; Cheng, Q.; Liu, Z. Immune Infiltrating Cells-Derived Risk Signature Based on Large-Scale Analysis Defines Immune Landscape and Predicts Immunotherapy Responses in Glioma Tumor Microenvironment. Front. Immunol. 2021, 1, 691811. [Google Scholar] [CrossRef] [PubMed]
- Han, X.J.; Ma, X.L.; Yang, L.; Wei, Y.Q.; Peng, Y.; Wei, X.W. Progress in Neoantigen Targeted Cancer Immunotherapies. Front. Cell Dev. Biol. 2020, 1, 728. [Google Scholar] [CrossRef] [PubMed]
- Blass, E.; Ott, P.A. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 2021, 1, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Stagg, J.; Smyth, M.J. Extracellular adenosine triphosphate and adenosine in cancer. Oncogene 2010, 1, 5346–5358. [Google Scholar] [CrossRef] [PubMed]
- Blay, J.; White, T.D.; Hoskin, D.W. The extracellular fluid of solid carcinomas contains immunosuppressive concentrations of adenosine. Cancer Res. 1997, 1, 2602–2605. [Google Scholar]
- Allard, B.; Longhi, M.S.; Robson, S.C.; Stagg, J. The ectonucleotidases CD39 and CD73: Novel checkpoint inhibitor targets. Immunol. Rev. 2017, 1, 121–144. [Google Scholar] [CrossRef] [Green Version]
- Cekic, C.; Linden, J. Purinergic regulation of the immune system. Nat. Rev. Immunol. 2016, 1, 177–192. [Google Scholar] [CrossRef]
- Tsukui, H.; Horie, H.; Koinuma, K.; Ohzawa, H.; Sakuma, Y.; Hosoya, Y.; Yamaguchi, H.; Yoshimura, K.; Lefor, A.K.; Sata, N.; et al. CD73 blockade enhances the local and abscopal effects of radiotherapy in a murine rectal cancer model. BMC Cancer 2020, 1, 411. [Google Scholar] [CrossRef] [PubMed]
- Stagg, J.; Divisekera, U.; McLaughlin, N.; Sharkey, J.; Pommey, S.; Denoyer, D.; Dwyer, K.M.; Smyth, M.J. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2010, 1, 1547–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sek, K.; Molck, C.; Stewart, G.D.; Kats, L.; Darcy, P.K.; Beavis, P.A. Targeting Adenosine Receptor Signaling in Cancer Immunotherapy. Int. J. Mol. Sci. 2018, 19, 3837. [Google Scholar] [CrossRef] [Green Version]
- Leone, R.D.; Emens, L.A. Targeting adenosine for cancer immunotherapy. J. Immunother. Cancer 2018, 1, 57. [Google Scholar] [CrossRef] [Green Version]
- Loi, S.; Pommey, S.; Haibe-Kains, B.; Beavis, P.A.; Darcy, P.K.; Smyth, M.J.; Stagg, J. CD73 promotes anthracycline resistance and poor prognosis in triple negative breast cancer. Proc. Natl. Acad. Sci. USA 2013, 1, 11091–11096. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.X.; Chen, Y.T.; Feng, B.; Mao, X.B.; Yu, B.; Chu, X.Y. Expression and clinical significance of CD73 and hypoxia-inducible factor-1alpha in gastric carcinoma. World J. Gastroenterol. 2013, 1, 1912–1918. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, S.; Floros, T.; Theodoraki, M.N.; Hong, C.S.; Jackson, E.K.; Lang, S.; Whiteside, T.L. Suppression of Lymphocyte Functions by Plasma Exosomes Correlates with Disease Activity in Patients with Head and Neck Cancer. Clin. Cancer Res. 2017, 1, 4843–4854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayan, D.; Young, A.; Teng, M.W.L.; Smyth, M.J. Targeting immunosuppressive adenosine in cancer. Nat. Rev. Cancer 2017, 1, 765. [Google Scholar] [CrossRef] [PubMed]
- Synnestvedt, K.; Furuta, G.T.; Comerford, K.M.; Louis, N.; Karhausen, J.; Eltzschig, H.K.; Hansen, K.R.; Thompson, L.F.; Colgan, S.P. Ecto-5′-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J. Clin. Investig. 2002, 1, 993–1002. [Google Scholar] [CrossRef]
- Jadidi-Niaragh, F. Potential of CD73 as a target for cancer immunotherapy. Immunotherapy 2019, 1, 1353–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghalamfarsa, G.; Kazemi, M.H.; Raoofi Mohseni, S.; Masjedi, A.; Hojjat-Farsangi, M.; Azizi, G.; Yousefi, M.; Jadidi-Niaragh, F. CD73 as a potential opportunity for cancer immunotherapy. Expert Opin. Ther. Targets 2019, 1, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Wang, Z.; Dai, Z.; Zhang, H.; Cheng, Q.; Liu, Z. Promoting Prognostic Model Application: A Review Based on Gliomas. J. Oncol. 2021, 1, 7840007. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; He, J.; Dai, Z.; Wang, Z.; Liang, X.; He, F.; Xia, Z.; Feng, S.; Cao, H.; Zhang, L.; et al. PDIA5 is Correlated with Immune Infiltration and Predicts Poor Prognosis in Gliomas. Front. Immunol. 2021, 1, 628966. [Google Scholar] [CrossRef]
- Farkona, S.; Diamandis, E.P.; Blasutig, I.M. Cancer immunotherapy: The beginning of the end of cancer? BMC Med. 2016, 1, 73. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Huang, J.; Liu, X.; Cheng, Q.; Luo, C.; Liu, Z. CTLA-4 correlates with immune and clinical characteristics of glioma. Cancer Cell Int. 2020, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- Lazzari, C.; Karachaliou, N.; Bulotta, A.; Vigano, M.; Mirabile, A.; Brioschi, E.; Santarpia, M.; Gianni, L.; Rosell, R.; Gregorc, V. Combination of immunotherapy with chemotherapy and radiotherapy in lung cancer: Is this the beginning of the end for cancer? Ther. Adv. Med. Oncol. 2018, 1, 1758835918762094. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 1, 76. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat. Immunol. 2010, 1, 889–896. [Google Scholar] [CrossRef]
- Zhao, H.; Garton, T.; Keep, R.F.; Hua, Y.; Xi, G. Microglia/Macrophage Polarization after Experimental Intracerebral Hemorrhage. Transl. Stroke Res. 2015, 1, 407–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasko, G.; Cronstein, B. Regulation of inflammation by adenosine. Front. Immunol. 2013, 1, 85. [Google Scholar] [CrossRef] [Green Version]
- Csoka, B.; Selmeczy, Z.; Koscso, B.; Nemeth, Z.H.; Pacher, P.; Murray, P.J.; Kepka-Lenhart, D.; Morris, S.M., Jr.; Gause, W.C.; Leibovich, S.J.; et al. Adenosine promotes alternative macrophage activation via A2A and A2B receptors. FASEB J. 2012, 1, 376–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 1, 651–668. [Google Scholar] [CrossRef]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 1, 116. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Sakaguchi, S. Targeting Treg cells in cancer immunotherapy. Eur. J. Immunol. 2019, 1, 1140–1146. [Google Scholar] [CrossRef] [Green Version]
- Ping, Q.; Yan, R.; Cheng, X.; Wang, W.; Zhong, Y.; Hou, Z.; Shi, Y.; Wang, C.; Li, R. Cancer-associated fibroblasts: Overview, progress, challenges, and directions. Cancer Gene Ther. 2021, 1, 984–999. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 1, 174–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Liu, W.; Yang, H.; Zhang, P.; Xiao, C.; Chen, X. A glutathione-responsive sulfur dioxide polymer prodrug as a nanocarrier for combating drug-resistance in cancer chemotherapy. Biomaterials 2018, 1, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 1, 1769–1792. [Google Scholar] [CrossRef]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, K.; Zhang, J.; Cao, H.; Xiao, G.; Wang, Z.; Zhang, X.; Zhang, N.; Wu, W.; Zhang, H.; Wang, Q.; et al. Identification of CD73 as a Novel Biomarker Encompassing the Tumor Microenvironment, Prognosis, and Therapeutic Responses in Various Cancers. Cancers 2022, 14, 5663. https://doi.org/10.3390/cancers14225663
Tang K, Zhang J, Cao H, Xiao G, Wang Z, Zhang X, Zhang N, Wu W, Zhang H, Wang Q, et al. Identification of CD73 as a Novel Biomarker Encompassing the Tumor Microenvironment, Prognosis, and Therapeutic Responses in Various Cancers. Cancers. 2022; 14(22):5663. https://doi.org/10.3390/cancers14225663
Chicago/Turabian StyleTang, Kun, Jingwei Zhang, Hui Cao, Gelei Xiao, Zeyu Wang, Xun Zhang, Nan Zhang, Wantao Wu, Hao Zhang, Qianrong Wang, and et al. 2022. "Identification of CD73 as a Novel Biomarker Encompassing the Tumor Microenvironment, Prognosis, and Therapeutic Responses in Various Cancers" Cancers 14, no. 22: 5663. https://doi.org/10.3390/cancers14225663
APA StyleTang, K., Zhang, J., Cao, H., Xiao, G., Wang, Z., Zhang, X., Zhang, N., Wu, W., Zhang, H., Wang, Q., Xu, H., & Cheng, Q. (2022). Identification of CD73 as a Novel Biomarker Encompassing the Tumor Microenvironment, Prognosis, and Therapeutic Responses in Various Cancers. Cancers, 14(22), 5663. https://doi.org/10.3390/cancers14225663