Differential Effects on the Translation of Immune-Related Alternatively Polyadenylated mRNAs in Melanoma and T Cells by eIF4A Inhibition
Abstract
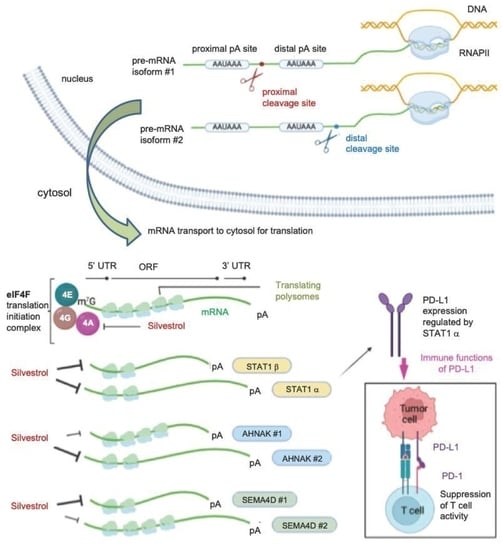
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and siRNA Transfections
2.2. Flow Cytometry Analysis
2.3. Western Blot
2.4. Polysomal Fractionation and Profiling
2.5. mRNA Preparation and Real-Time/Quantitative PCR
2.6. 3′-Seq Experiments
2.7. 3′-Seq Bioinformatic Analysis
2.8. Statistics
3. Results
3.1. Functional Importance of APA-Generated STAT1 Protein Isoforms for PD-L1 Gene Expression
3.2. Both STAT1 mRNA Isoforms Are Regulated by eIF4A Inhibition at the Translational Level
3.3. eIF4A Inhibition Regulates the Translation of mRNAs Encoding Key Immune Checkpoint Proteins in Activated T Cells
3.4. eIF4A Inhibition Differentially Regulates the Translation of APA Isoforms in Several Immune-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Restifo, N.P.; Smyth, M.J.; Snyder, A. Acquired Resistance to Immunotherapy and Future Challenges. Nat. Rev. Cancer 2016, 16, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budhwani, M.; Turrell, G.; Yu, M.; Frazer, I.H.; Mehdi, A.M.; Chandra, J. Immune-Inhibitory Gene Expression Is Positively Correlated with Overall Immune Activity and Predicts Increased Survival Probability of Cervical and Head and Neck Cancer Patients. Front. Mol. Biosci. 2021, 8, 622643. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, S.; Meller, J.; Hongeng, S.; Tohtong, R.; Chutipongtanate, S. Transcriptional Regulation of Cancer Immune Checkpoints: Emerging Strategies for Immunotherapy. Vaccines 2020, 8, 735. [Google Scholar] [CrossRef]
- Xu, H.-H.; Gan, J.; Xu, D.-P.; Li, L.; Yan, W.-H. Comprehensive Transcriptomic Analysis Reveals the Role of the Immune Checkpoint HLA-G Molecule in Cancers. Front. Immunol. 2021, 12, 614773. [Google Scholar] [CrossRef]
- Zerdes, I.; Matikas, A.; Bergh, J.; Rassidakis, G.Z.; Foukakis, T. Genetic, Transcriptional and Post-Translational Regulation of the Programmed Death Protein Ligand 1 in Cancer: Biology and Clinical Correlations. Oncogene 2018, 37, 4639–4661. [Google Scholar] [CrossRef] [Green Version]
- Neve, J.; Patel, R.; Wang, Z.; Louey, A.; Furger, A.M. Cleavage and Polyadenylation: Ending the Message Expands Gene Regulation. RNA Biol. 2017, 14, 865–890. [Google Scholar] [CrossRef] [Green Version]
- Passmore, L.A.; Tang, T.T. The Long and Short of It. eLife 2021, 10, e70757. [Google Scholar] [CrossRef]
- Tian, B.; Graber, J.H. Signals for Pre-MRNA Cleavage and Polyadenylation: Polyadenylation Signals. Wiley Interdiscip. Rev. RNA 2012, 3, 385–396. [Google Scholar] [CrossRef] [Green Version]
- Xiang, K.; Bartel, D.P. The Molecular Basis of Coupling between Poly(A)-Tail Length and Translational Efficiency. eLife 2021, 10, e66493. [Google Scholar] [CrossRef]
- Derti, A.; Garrett-Engele, P.; MacIsaac, K.D.; Stevens, R.C.; Sriram, S.; Chen, R.; Rohl, C.A.; Johnson, J.M.; Babak, T. A Quantitative Atlas of Polyadenylation in Five Mammals. Genome Res. 2012, 22, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of Post-Transcriptional Regulation by MicroRNAs: Are the Answers in Sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Rehfeld, A.; Plass, M.; Krogh, A.; Friis-Hansen, L. Alterations in Polyadenylation and Its Implications for Endocrine Disease. Front. Endocrinol. 2013, 4, 53. [Google Scholar] [CrossRef] [Green Version]
- Zanzoni, A.; Spinelli, L.; Ribeiro, D.M.; Tartaglia, G.G.; Brun, C. Post-Transcriptional Regulatory Patterns Revealed by Protein-RNA Interactions. Sci. Rep. 2019, 9, 4302. [Google Scholar] [CrossRef] [Green Version]
- Boussemart, L.; Malka-Mahieu, H.; Girault, I.; Allard, D.; Hemmingsson, O.; Tomasic, G.; Thomas, M.; Basmadjian, C.; Ribeiro, N.; Thuaud, F.; et al. EIF4F Is a Nexus of Resistance to Anti-BRAF and Anti-MEK Cancer Therapies. Nature 2014, 513, 105–109. [Google Scholar] [CrossRef]
- Cerezo, M.; Guemiri, R.; Druillennec, S.; Girault, I.; Malka-Mahieu, H.; Shen, S.; Allard, D.; Martineau, S.; Welsch, C.; Agoussi, S.; et al. Translational Control of Tumor Immune Escape via the EIF4F–STAT1–PD-L1 Axis in Melanoma. Nat. Med. 2018, 24, 1877–1886. [Google Scholar] [CrossRef]
- Malka-Mahieu, H.; Girault, I.; Rubington, M.; Leriche, M.; Welsch, C.; Kamsu-Kom, N.; Zhao, Q.; Desaubry, L.; Vagner, S.; Robert, C. Synergistic Effects of EIF4A and MEK Inhibitors on Proliferation of NRAS-Mutant Melanoma Cell Lines. Cell Cycle 2016, 15, 2405–2409. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.; Faouzi, S.; Bastide, A.; Martineau, S.; Malka-Mahieu, H.; Fu, Y.; Sun, X.; Mateus, C.; Routier, E.; Roy, S.; et al. An Epitranscriptomic Mechanism Underlies Selective MRNA Translation Remodelling in Melanoma Persister Cells. Nat. Commun. 2019, 10, 5713. [Google Scholar] [CrossRef] [Green Version]
- Fabbri, L.; Chakraborty, A.; Robert, C.; Vagner, S. The Plasticity of MRNA Translation during Cancer Progression and Therapy Resistance. Nat. Rev. Cancer 2021, 21, 558–577. [Google Scholar] [CrossRef]
- Bordeleau, M.-E.; Robert, F.; Gerard, B.; Lindqvist, L.; Chen, S.M.H.; Wendel, H.-G.; Brem, B.; Greger, H.; Lowe, S.W.; Porco, J.A.; et al. Therapeutic Suppression of Translation Initiation Modulates Chemosensitivity in a Mouse Lymphoma Model. J. Clin. Investig. 2008, 118, 2651–2660. [Google Scholar] [CrossRef] [Green Version]
- Chu, J.; Galicia-Vázquez, G.; Cencic, R.; Mills, J.R.; Katigbak, A.; Porco, J.A.; Pelletier, J. CRISPR-Mediated Drug-Target Validation Reveals Selective Pharmacological Inhibition of the RNA Helicase, EIF4A. Cell Rep. 2016, 15, 2340–2347. [Google Scholar] [CrossRef] [Green Version]
- Sadlish, H.; Galicia-Vazquez, G.; Paris, C.G.; Aust, T.; Bhullar, B.; Chang, L.; Helliwell, S.B.; Hoepfner, D.; Knapp, B.; Riedl, R.; et al. Evidence for a Functionally Relevant Rocaglamide Binding Site on the EIF4A–RNA Complex. ACS Chem. Biol. 2013, 8, 1519–1527. [Google Scholar] [CrossRef] [Green Version]
- Cencic, R.; Carrier, M.; Galicia-Vázquez, G.; Bordeleau, M.-E.; Sukarieh, R.; Bourdeau, A.; Brem, B.; Teodoro, J.G.; Greger, H.; Tremblay, M.L.; et al. Antitumor Activity and Mechanism of Action of the Cyclopenta[b]Benzofuran, Silvestrol. PLoS ONE 2009, 4, e5223. [Google Scholar] [CrossRef] [Green Version]
- Kogure, T.; Kinghorn, A.D.; Yan, I.; Bolon, B.; Lucas, D.M.; Grever, M.R.; Patel, T. Therapeutic Potential of the Translation Inhibitor Silvestrol in Hepatocellular Cancer. PLoS ONE 2013, 8, e76136. [Google Scholar] [CrossRef]
- Rubio, C.A.; Weisburd, B.; Holderfield, M.; Arias, C.; Fang, E.; DeRisi, J.L.; Fanidi, A. Transcriptome-Wide Characterization of the EIF4A Signature Highlights Plasticity in Translation Regulation. Genome Biol. 2014, 15, 476. [Google Scholar] [CrossRef]
- Schulz, G.; Victoria, C.; Kirschning, A.; Steinmann, E. Rocaglamide and Silvestrol: A Long Story from Anti-Tumor to Anti-Coronavirus Compounds. Nat. Prod. Rep. 2021, 38, 18–23. [Google Scholar] [CrossRef]
- Singh, I.; Lee, S.-H.; Sperling, A.S.; Samur, M.K.; Tai, Y.-T.; Fulciniti, M.; Munshi, N.C.; Mayr, C.; Leslie, C.S. Widespread Intronic Polyadenylation Diversifies Immune Cell Transcriptomes. Nat. Commun. 2018, 9, 1716. [Google Scholar] [CrossRef]
- Liu, S.; Kang, W.-J.; Abrimian, A.; Xu, J.; Cartegni, L.; Majumdar, S.; Hesketh, P.; Bekker, A.; Pan, Y.-X. Alternative Pre-MRNA Splicing of the Mu Opioid Receptor Gene, OPRM1: Insight into Complex Mu Opioid Actions. Biomolecules 2021, 11, 1525. [Google Scholar] [CrossRef]
- Vorlová, S.; Rocco, G.; LeFave, C.V.; Jodelka, F.M.; Hess, K.; Hastings, M.L.; Henke, E.; Cartegni, L. Induction of Antagonistic Soluble Decoy Receptor Tyrosine Kinases by Intronic PolyA Activation. Mol. Cell 2011, 43, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Zammarchi, F.; Boutsalis, G.; Cartegni, L. 5′ UTR Control of Native ERG and of Tmprss2:ERG Variants Activity in Prostate Cancer. PLoS ONE 2013, 8, e49721. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- GitHub. Available online: https://github.com/InstitutCurie/3-SMART (accessed on 19 January 2022).
- Herrmann, C.J.; Schmidt, R.; Kanitz, A.; Artimo, P.; Gruber, A.J.; Zavolan, M. PolyASite 2.0: A Consolidated Atlas of Polyadenylation Sites from 3′ End Sequencing. Nucleic Acids Res. 2020, 48, D174–D179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Zheng, D.; Yehia, G.; Tian, B. A Compendium of Conserved Cleavage and Polyadenylation Events in Mammalian Genes. Genome Res. 2018, 28, 1427–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittendorf, E.A.; Philips, A.V.; Meric-Bernstam, F.; Qiao, N.; Wu, Y.; Harrington, S.; Su, X.; Wang, Y.; Gonzalez-Angulo, A.M.; Akcakanat, A.; et al. PD-L1 Expression in Triple-Negative Breast Cancer. Cancer Immunol. Res. 2014, 2, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Matza, D.; Badou, A.; Jha, M.K.; Willinger, T.; Antov, A.; Sanjabi, S.; Kobayashi, K.S.; Marchesi, V.T.; Flavell, R.A. Requirement for AHNAK1-Mediated Calcium Signaling during T Lymphocyte Cytolysis. Proc. Natl. Acad. Sci. USA 2009, 106, 9785–9790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matza, D.; Badou, A.; Kobayashi, K.S.; Goldsmith-Pestana, K.; Masuda, Y.; Komuro, A.; McMahon-Pratt, D.; Marchesi, V.T.; Flavell, R.A. A Scaffold Protein, AHNAK1, Is Required for Calcium Signaling during T Cell Activation. Immunity 2008, 28, 64–74. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Fang, Y.; Zhai, B.; Liu, X.; Zhu, G.; Zhou, S.; Xu, Y.; Wang, X.; Su, W.; Wang, R. Gm40600 Promotes CD4 + T-cell Responses by Interacting with Ahnak. Immunology 2021, 164, 190–206. [Google Scholar] [CrossRef]
- Kim, I.Y.; Yi, S.S.; Shin, J.H.; Kim, Y.N.; Ko, C.-Y.; Kim, H.S.; Lee, S.Y.; Bae, Y.S.; Seong, J.K. Intensive Morphometric Analysis of Enormous Alterations in Skeletal Bone System with Micro-CT for AHNAK−/− Mice. Anat. Sci. Int. 2020, 95, 323–333. [Google Scholar] [CrossRef]
- Choi, E.W.; Lee, H.W.; Lee, J.S.; Kim, I.Y.; Shin, J.H.; Seong, J.K. Ahnak-Knockout Mice Show Susceptibility to Bartonella Henselae Infection Because of CD4+ T Cell Inactivation and Decreased Cytokine Secretion. BMB Rep. 2019, 52, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Björkström, N.K.; Melum, E. Intact CD100–CD72 Interaction Necessary for TCR-Induced T Cell Proliferation. Front. Immunol. 2017, 8, 765. [Google Scholar] [CrossRef] [Green Version]
- Kuklina, E.; Nekrasova, I.; Glebezdina, N. Signaling from Membrane Semaphorin 4D in T Lymphocytes. Mol. Immunol. 2021, 129, 56–62. [Google Scholar] [CrossRef]
- Xie, J.; Wang, Z.; Wang, W. Semaphorin 4D Induces an Imbalance of Th17/Treg Cells by Activating the Aryl Hydrocarbon Receptor in Ankylosing Spondylitis. Front. Immunol. 2020, 11, 2151. [Google Scholar] [CrossRef]
- Evans, E.E.; Jonason, A.S.; Bussler, H.; Torno, S.; Veeraraghavan, J.; Reilly, C.; Doherty, M.A.; Seils, J.; Winter, L.A.; Mallow, C.; et al. Antibody Blockade of Semaphorin 4D Promotes Immune Infiltration into Tumor and Enhances Response to Other Immunomodulatory Therapies. Cancer Immunol. Res. 2015, 3, 689–701. [Google Scholar] [CrossRef] [Green Version]
- Rossi, A.J.; Khan, T.M.; Hong, H.; Lesinski, G.B.; Wu, C.; Hernandez, J.M. Pepinemab (Anti-SEMA4D) in Combination with Ipilimumab or Nivolumab for Patients with Resectable Pancreatic and Colorectal Cancer. Ann. Surg. Oncol. 2021, 28, 4098–4099. [Google Scholar] [CrossRef]
- Domingues, R.G.; Lago-Baldaia, I.; Pereira-Castro, I.; Fachini, J.M.; Oliveira, L.; Drpic, D.; Lopes, N.; Henriques, T.; Neilson, J.R.; Carmo, A.M.; et al. CD5 Expression Is Regulated during Human T-Cell Activation by Alternative Polyadenylation, PTBP1, and MiR-204. Eur. J. Immunol. 2016, 46, 1490–1503. [Google Scholar] [CrossRef] [Green Version]
- Beisang, D.; Reilly, C.; Bohjanen, P.R. Alternative Polyadenylation Regulates CELF1/CUGBP1 Target Transcripts Following T Cell Activation. Gene 2014, 550, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Gruber, A.R.; Martin, G.; Müller, P.; Schmidt, A.; Gruber, A.J.; Gumienny, R.; Mittal, N.; Jayachandran, R.; Pieters, J.; Keller, W.; et al. Global 3′ UTR Shortening Has a Limited Effect on Protein Abundance in Proliferating T Cells. Nat. Commun. 2014, 5, 5465. [Google Scholar] [CrossRef] [Green Version]
- Sandberg, R.; Neilson, J.R.; Sarma, A.; Sharp, P.A.; Burge, C.B. Proliferating Cells Express MRNAs with Shortened 3′ Untranslated Regions and Fewer MicroRNA Target Sites. Science 2008, 320, 1643–1647. [Google Scholar] [CrossRef] [Green Version]
- Spies, N.; Burge, C.B.; Bartel, D.P. 3′ UTR-Isoform Choice Has Limited Influence on the Stability and Translational Efficiency of Most MRNAs in Mouse Fibroblasts. Genome Res. 2013, 23, 2078–2090. [Google Scholar] [CrossRef] [Green Version]
- Waldron, J.A.; Tack, D.C.; Ritchey, L.E.; Gillen, S.L.; Wilczynska, A.; Turro, E.; Bevilacqua, P.C.; Assmann, S.M.; Bushell, M.; Le Quesne, J. MRNA Structural Elements Immediately Upstream of the Start Codon Dictate Dependence upon EIF4A Helicase Activity. Genome Biol. 2019, 20, 300. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van der Meulen, J.; et al. RNA G-Quadruplexes Cause EIF4A-Dependent Oncogene Translation in Cancer. Nature 2014, 513, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Christensen, A.K.; Kahn, L.E.; Bourne, C.M. Circular Polysomes Predominate on the Rough Endoplasmic Reticulum of Somatotropes and Mammotropes in the Rat Anterior Pituitary. Am. J. Anat. 1987, 178, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Christensen, A.K.; Bourne, C.M. Shape of Large Bound Polysomes in Cultured Fibroblasts and Thyroid Epithelial Cells. Anat. Rec. 1999, 255, 116–129. [Google Scholar] [CrossRef] [Green Version]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. MRNA Splicing-Mediated C-Terminal Replacement of Transcription Factor Hac1p Is Required for Efficient Activation of the Unfolded Protein Response. Proc. Natl. Acad. Sci. USA 2000, 97, 4660–4665. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Mayr, C. A Membraneless Organelle Associated with the Endoplasmic Reticulum Enables 3′UTR-Mediated Protein-Protein Interactions. Cell 2018, 175, 1492–1506.e19. [Google Scholar] [CrossRef] [Green Version]
- Khong, A.; Matheny, T.; Jain, S.; Mitchell, S.F.; Wheeler, J.R.; Parker, R. The Stress Granule Transcriptome Reveals Principles of MRNA Accumulation in Stress Granules. Mol. Cell 2017, 68, 808–820.e5. [Google Scholar] [CrossRef] [PubMed]
- Namkoong, S.; Ho, A.; Woo, Y.M.; Kwak, H.; Lee, J.H. Systematic Characterization of Stress-Induced RNA Granulation. Mol. Cell 2018, 70, 175–187.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franchini, D.-M.; Lanvin, O.; Tosolini, M.; Patras de Campaigno, E.; Cammas, A.; Péricart, S.; Scarlata, C.-M.; Lebras, M.; Rossi, C.; Ligat, L.; et al. Microtubule-Driven Stress Granule Dynamics Regulate Inhibitory Immune Checkpoint Expression in T Cells. Cell Rep. 2019, 26, 94–107.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, C. LAG-3 and PD-1 Blockade Raises the Bar for Melanoma. Nat. Cancer 2021, 2, 1251–1253. [Google Scholar] [CrossRef]
- Acharya, N.; Sabatos-Peyton, C.; Anderson, A.C. Tim-3 Finds Its Place in the Cancer Immunotherapy Landscape. J. Immunother. Cancer 2020, 8, e000911. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswas, B.; Guemiri, R.; Cadix, M.; Labbé, C.M.; Chakraborty, A.; Dutertre, M.; Robert, C.; Vagner, S. Differential Effects on the Translation of Immune-Related Alternatively Polyadenylated mRNAs in Melanoma and T Cells by eIF4A Inhibition. Cancers 2022, 14, 1177. https://doi.org/10.3390/cancers14051177
Biswas B, Guemiri R, Cadix M, Labbé CM, Chakraborty A, Dutertre M, Robert C, Vagner S. Differential Effects on the Translation of Immune-Related Alternatively Polyadenylated mRNAs in Melanoma and T Cells by eIF4A Inhibition. Cancers. 2022; 14(5):1177. https://doi.org/10.3390/cancers14051177
Chicago/Turabian StyleBiswas, Biswendu, Ramdane Guemiri, Mandy Cadix, Céline M. Labbé, Alina Chakraborty, Martin Dutertre, Caroline Robert, and Stéphan Vagner. 2022. "Differential Effects on the Translation of Immune-Related Alternatively Polyadenylated mRNAs in Melanoma and T Cells by eIF4A Inhibition" Cancers 14, no. 5: 1177. https://doi.org/10.3390/cancers14051177
APA StyleBiswas, B., Guemiri, R., Cadix, M., Labbé, C. M., Chakraborty, A., Dutertre, M., Robert, C., & Vagner, S. (2022). Differential Effects on the Translation of Immune-Related Alternatively Polyadenylated mRNAs in Melanoma and T Cells by eIF4A Inhibition. Cancers, 14(5), 1177. https://doi.org/10.3390/cancers14051177