Target Genes of c-MYC and MYCN with Prognostic Power in Neuroblastoma Exhibit Different Expressions during Sympathoadrenal Development
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. c-MYC/MYCN Target-Gene Sets
2.2. Expression Analysis of NB Cohorts
2.3. Analysis of the Prognostic Value of c-MYC/MYCN Target Genes in NB

2.4. Prognostic Power Evaluation by Survival and Efficiency Analysis
2.5. Cross-Species Comparisons and Gene Ontology Analysis
2.6. Single Cell Analysis of Gene Expression Profiles during Sympathoadrenal Development and NB
2.7. Statistical Analysis
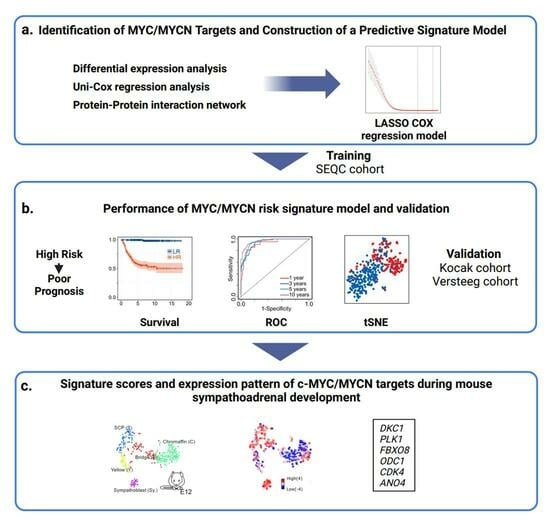
3. Results
3.1. Development of an Analytical Framework to Screen c-MYC/MYCN Target Genes with Prognostic Value in NB
3.2. Expression of c-MYC/MYCN Target Genes Stratify NB Patients in Clinical Groups with Different Outcomes
3.3. c-MYC/MYCN Prognostic Target Genes Are Direct Targets of c-MYC/MYCN in NB
3.4. Comparative Analysis of Patient Risk Stratification between c-MYC/MYCN Target Genes and Markers of Sympathoadrenal Development and NB
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; Thompson, T.D.; Miller, J.W.; Pollack, L.A.; Stewart, S.L. Cancer incidence among children and adolescents in the United States, 2001–2003. Pediatrics 2008, 121, e1470–e1477. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.H.; Meng, M.V.; Walsh, T.J.; Matthay, K.K.; Baskin, L.S. Increasing incidence of neuroblastoma and potentially higher associated mortality of children from nonmetropolitan areas: Analysis of the surveillance, epidemiology, and end results database. J. Pediatr. Hematol. Oncol. 2009, 31, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Primers 2016, 2, 16078. [Google Scholar] [CrossRef]
- Newman, E.A.; Abdessalam, S.; Aldrink, J.H.; Austin, M.; Heaton, T.E.; Bruny, J.; Ehrlich, P.; Dasgupta, R.; Baertschiger, R.M.; Lautz, T.B.; et al. Update on neuroblastoma. J. Pediatr. Surg. 2019, 54, 383–389. [Google Scholar] [CrossRef]
- Cheung, N.K.; Dyer, M.A. Neuroblastoma: Developmental biology, cancer genomics and immunotherapy. Nat. Rev. Cancer 2013, 13, 397–411. [Google Scholar] [CrossRef]
- Grobner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef]
- Bown, N.; Cotterill, S.; Lastowska, M.; O’Neill, S.; Pearson, A.D.; Plantaz, D.; Meddeb, M.; Danglot, G.; Brinkschmidt, C.; Christiansen, H.; et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N. Engl. J. Med. 1999, 340, 1954–1961. [Google Scholar] [CrossRef]
- Lastowska, M.; Viprey, V.; Santibanez-Koref, M.; Wappler, I.; Peters, H.; Cullinane, C.; Roberts, P.; Hall, A.G.; Tweddle, D.A.; Pearson, A.D.; et al. Identification of candidate genes involved in neuroblastoma progression by combining genomic and expression microarrays with survival data. Oncogene 2007, 26, 7432–7444. [Google Scholar] [CrossRef]
- Attiyeh, E.F.; London, W.B.; Mosse, Y.P.; Wang, Q.; Winter, C.; Khazi, D.; McGrady, P.W.; Seeger, R.C.; Look, A.T.; Shimada, H.; et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 2005, 353, 2243–2253. [Google Scholar] [CrossRef]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.; Castel, V.; Castelberry, R.P.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F.; et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar] [CrossRef]
- Park, J.R.; Bagatell, R.; London, W.B.; Maris, J.M.; Cohn, S.L.; Mattay, K.K.; Hogarty, M.; COG Neuroblastoma Committee. Children’s Oncology Group’s 2013 blueprInt. for research: Neuroblastoma. Pediatr. Blood Cancer 2013, 60, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Irwin, M.S.; Naranjo, A.; Zhang, F.F.; Cohn, S.L.; London, W.B.; Gastier-Foster, J.M.; Ramirez, N.C.; Pfau, R.; Reshmi, S.; Wagner, E.; et al. Revised neuroblastoma risk classification system: A report from the Children’s Oncology Group. J. Clin. Oncol. 2021, 39, 3229–3241. [Google Scholar] [CrossRef] [PubMed]
- Cohn, S.L.; Pearson, A.D.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The international Neuroblastoma Risk Group (INRG) classification system: An INRG task force report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef]
- Pinto, N.; Naranjo, A.; Hibbitts, E.; Kreissman, S.G.; Granger, M.M.; Irwin, M.S.; Bagatell, R.; London, W.B.; Greengard, E.G.; Park, J.R.; et al. Predictors of differential response to induction therapy in high-risk neuroblastoma: A report from the Children’s Oncology Group (COG). Eur. J. Cancer 2019, 112, 66–79. [Google Scholar] [CrossRef]
- Liang, W.H.; Federico, S.M.; London, W.B.; Naranjo, A.; Irwin, M.S.; Volchenboum, S.L.; Cohn, S.L. Tailoring therapy for children with neuroblastoma on the basis of risk group classification: Past, present, and future. JCO Clin. Cancer Inform. 2020, 4, 895–905. [Google Scholar] [CrossRef]
- Nader, J.H.; Bourgeois, F.; Bagatell, R.; Moreno, L.; Pearson, A.D.J.; DuBois, S.G. Systematic review of clinical drug development activities for neuroblastoma from 2011 to 2020. Pediatr. Blood Cancer 2023, 70, e30106. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Seeger, R.C.; Schwab, M.; Varmus, H.E.; Bishop, J.M. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984, 224, 1121–1124. [Google Scholar] [CrossRef]
- Otte, J.; Dyberg, C.; Pepich, A.; Johnsen, J.I. MYCN function in neuroblastoma development. Front. Oncol. 2020, 10, 624079. [Google Scholar] [CrossRef]
- Vennstrom, B.; Sheiness, D.; Zabielski, J.; Bishop, J.M. Isolation and characterization of c-myc, a cellular homolog of the oncogene (v-myc) of avian myelocytomatosis virus strain 29. J. Virol. 1982, 42, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Seeger, R.C.; Brodeur, G.M.; Sather, H.; Dalton, A.; Siegel, S.E.; Wong, K.Y.; Hammond, D. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N. Engl. J. Med. 1985, 313, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Wolfer, A.; Ramaswamy, S. MYC and metastasis. Cancer Res. 2011, 71, 2034–2037. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef]
- Rickman, D.S.; Schulte, J.H.; Eilers, M. The expanding world of N-MYC-driven tumors. Cancer Discov. 2018, 8, 150–163. [Google Scholar] [CrossRef]
- Paglia, S.; Sollazzo, M.; Di Giacomo, S.; Strocchi, S.; Grifoni, D. Exploring MYC relevance to cancer biology from the perspective of cell competition. SeMin. Cancer Biol. 2020, 63, 49–59. [Google Scholar] [CrossRef]
- Madden, S.K.; de Araujo, A.D.; Gerhardt, M.; Fairlie, D.P.; Mason, J.M. Taking the Myc out of cancer: Toward therapeutic strategies to directly inhibit c-Myc. Mol. Cancer 2021, 20, 3. [Google Scholar] [CrossRef]
- Huang, M.; Weiss, W.A. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 2013, 3, a014415. [Google Scholar] [CrossRef]
- Zimmerman, K.A.; Yancopoulos, G.D.; Collum, R.G.; Smith, R.K.; Kohl, N.E.; Denis, K.A.; Nau, M.M.; Witte, O.N.; Toran-Allerand, D.; Gee, C.E.; et al. Differential expression of myc family genes during murine development. Nature 1986, 319, 780–783. [Google Scholar] [CrossRef]
- Downs, K.M.; Martin, G.R.; Bishop, J.M. Contrasting patterns of myc and N-myc expression during gastrulation of the mouse embryo. Genes Dev. 1989, 3, 860–869. [Google Scholar] [CrossRef]
- DePinho, R.A.; Schreiber-Agus, N.; Alt, F.W. myc family oncogenes in the development of normal and neoplastic cells. Adv. Cancer Res. 1991, 57, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Weiss, W.A.; Aldape, K.; Mohapatra, G.; Feuerstein, B.G.; Bishop, J.M. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997, 16, 2985–2995. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yeo, K.S.; Levee, T.M.; Howe, C.J.; Her, Z.P.; Zhu, S. Zebrafish as a neuroblastoma model: Progress made, promise for the future. Cells 2021, 10, 580. [Google Scholar] [CrossRef]
- Nakagawa, M.; Takizawa, N.; Narita, M.; Ichisaka, T.; Yamanaka, S. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. USA 2010, 107, 14152–14157. [Google Scholar] [CrossRef]
- Varlakhanova, N.V.; Cotterman, R.F.; deVries, W.N.; Morgan, J.; Donahue, L.R.; Murray, S.; Knowles, B.B.; Knoepfler, P.S. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation 2010, 80, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Chappell, J.; Dalton, S. Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb. Perspect. Med. 2013, 3, a014381. [Google Scholar] [CrossRef]
- Westermann, F.; Muth, D.; Benner, A.; Bauer, T.; Henrich, K.O.; Oberthuer, A.; Brors, B.; Beissbarth, T.; Vandesompele, J.; Pattyn, F.; et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008, 9, R150. [Google Scholar] [CrossRef]
- Santarius, T.; Shipley, J.; Brewer, D.; Stratton, M.R.; Cooper, C.S. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 2010, 10, 59–64. [Google Scholar] [CrossRef]
- Schmitt-Hoffner, F.; van Rijn, S.; Toprak, U.H.; Mauermann, M.; Rosemann, F.; Heit-Mondrzyk, A.; Hubner, J.M.; Camgoz, A.; Hartlieb, S.; Pfister, S.M.; et al. FOXR2 stabilizes MYCN protein and identifies non-MYCN-amplified neuroblastoma patients with unfavorable outcome. J. Clin. Oncol. 2021, 39, 3217–3228. [Google Scholar] [CrossRef]
- Tang, X.X.; Zhao, H.; Kung, B.; Kim, D.Y.; Hicks, S.L.; Cohn, S.L.; Cheung, N.K.; Seeger, R.C.; Evans, A.E.; Ikegaki, N. The MYCN enigma: Significance of MYCN expression in neuroblastoma. Cancer Res. 2006, 66, 2826–2833. [Google Scholar] [CrossRef]
- Powers, J.T.; Tsanov, K.M.; Pearson, D.S.; Roels, F.; Spina, C.S.; Ebright, R.; Seligson, M.; de Soysa, Y.; Cahan, P.; Theissen, J.; et al. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 2016, 535, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, M.W.; Liu, Y.; He, S.; Durbin, A.D.; Abraham, B.J.; Easton, J.; Shao, Y.; Xu, B.; Zhu, S.; Zhang, X.; et al. MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov. 2018, 8, 320–335. [Google Scholar] [CrossRef] [PubMed]
- Valentijn, L.J.; Koster, J.; Haneveld, F.; Aissa, R.A.; van Sluis, P.; Broekmans, M.E.; Molenaar, J.J.; van Nes, J.; Versteeg, R. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc. Natl. Acad. Sci. USA 2012, 109, 19190–19195. [Google Scholar] [CrossRef]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular SignatuRes. Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.H.; Iyer, V.R. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS ONE 2008, 3, e1798. [Google Scholar] [CrossRef]
- Consortium, S.M.-I. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat. Biotechnol. 2014, 32, 903–914. [Google Scholar] [CrossRef]
- Su, Z.; Fang, H.; Hong, H.; Shi, L.; Zhang, W.; Zhang, W.; Zhang, Y.; Dong, Z.; Lancashire, L.J.; Bessarabova, M.; et al. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014, 15, 523. [Google Scholar] [CrossRef]
- Zhang, W.; Yu, Y.; Hertwig, F.; Thierry-Mieg, J.; Zhang, W.; Thierry-Mieg, D.; Wang, J.; Furlanello, C.; Devanarayan, V.; Cheng, J.; et al. Comparison of RNA-seq and microarray-based models for clinical endpoInt. prediction. Genome Biol. 2015, 16, 133. [Google Scholar] [CrossRef] [PubMed]
- Kocak, H.; Ackermann, S.; Hero, B.; Kahlert, Y.; Oberthuer, A.; Juraeva, D.; Roels, F.; Theissen, J.; Westermann, F.; Deubzer, H.; et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013, 4, e586. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Koster, J.; Zwijnenburg, D.A.; van Sluis, P.; Valentijn, L.J.; van der Ploeg, I.; Hamdi, M.; van Nes, J.; Westerman, B.A.; van Arkel, J.; et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012, 483, 589–593. [Google Scholar] [CrossRef]
- Oberthuer, A.; Juraeva, D.; Li, L.; Kahlert, Y.; Westermann, F.; Eils, R.; Berthold, F.; Shi, L.; Wolfinger, R.D.; Fischer, M.; et al. Comparison of performance of one-color and two-color gene-expression analyses in predicting clinical endpoints of neuroblastoma patients. Pharmacogenom. J. 2010, 10, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Benner, A.; Zucknick, M.; Hielscher, T.; Ittrich, C.; Mansmann, U. High-dimensional Cox models: The choice of penalty as part of the model building process. Biom. J. 2010, 52, 50–69. [Google Scholar] [CrossRef] [PubMed]
- Jardillier, R.; Koca, D.; Chatelain, F.; Guyon, L. Prognosis of lasso-like penalized Cox models with tumor profiling improves prediction over clinical data alone and benefits from bi-dimensional pre-screening. BMC Cancer 2022, 22, 1045. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 2018, 57, 289–300. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, X.; Peng, F.; Li, W.; Li, J.J. Exaggerated false positives by popular differential expression methods when analyzing human population samples. Genome Biol. 2022, 23, 79. [Google Scholar] [CrossRef]
- Heagerty, P.J.; Lumley, T.; Pepe, M.S. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000, 56, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Furlan, A.; Dyachuk, V.; Kastriti, M.E.; Calvo-Enrique, L.; Abdo, H.; Hadjab, S.; Chontorotzea, T.; Akkuratova, N.; Usoskin, D.; Kamenev, D.; et al. Multipotent peripheral glial cells generate neuroendocrine cells of the adrenal medulla. Science 2017, 357, eaal3753. [Google Scholar] [CrossRef] [PubMed]
- Jansky, S.; Sharma, A.K.; Korber, V.; Quintero, A.; Toprak, U.H.; Wecht, E.M.; Gartlgruber, M.; Greco, A.; Chomsky, E.; Grunewald, T.G.P.; et al. Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat. Genet. 2021, 53, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Van Groningen, T.; Koster, J.; Valentijn, L.J.; Zwijnenburg, D.A.; Akogul, N.; Hasselt, N.E.; Broekmans, M.; Haneveld, F.; Nowakowska, N.E.; Bras, J.; et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 2017, 49, 1261–1266. [Google Scholar] [CrossRef]
- Kildisiute, G.; Kholosy, W.M.; Young, M.D.; Roberts, K.; Elmentaite, R.; van Hooff, S.R.; Pacyna, C.N.; Khabirova, E.; Piapi, A.; Thevanesan, C.; et al. Tumor to normal single-cell mRNA comparisons reveal a pan-neuroblastoma cancer cell. Sci. Adv. 2021, 7, eabd3311. [Google Scholar] [CrossRef]
- Bedoya-Reina, O.C.; Li, W.; Arceo, M.; Plescher, M.; Bullova, P.; Pui, H.; Kaucka, M.; Kharchenko, P.; Martinsson, T.; Holmberg, J.; et al. Single-nuclei transcriptomes from human adrenal gland reveal distinct cellular identities of low and high-risk neuroblastoma tumors. Nat. Commun. 2021, 12, 5309. [Google Scholar] [CrossRef]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudie, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Zhou, L.; Tang, L.; Song, A.T.; Cibrik, D.M.; Song, P.X. A LASSO method to identify protein signature predicting post-transplant renal graft survival. Stat. Biosci. 2017, 9, 431–452. [Google Scholar] [CrossRef]
- Gbadamosi, M.O.; Shastri, V.M.; Elsayed, A.H.; Ries, R.; Olabige, O.; Nguyen, N.H.K.; De Jesus, A.; Wang, Y.C.; Dang, A.; Hirsch, B.A.; et al. A ten-gene DNA-damage response pathway gene expression signature predicts gemtuzumab ozogamicin response in pediatric AML patients treated on COGAAML0531 and AAML03P1 trials. Leukemia 2022, 36, 2022–2031. [Google Scholar] [CrossRef]
- Tu, H.; Zhang, Q.; Xue, L.; Bao, J. The cuproptosis-related lncRNA gene signature establishes a prognostic model of gastric adenocarcinoma and evaluates the effect of antineoplastic drugs. Genes 2022, 13, 2214. [Google Scholar] [CrossRef] [PubMed]
- Upton, K.; Modi, A.; Patel, K.; Kendsersky, N.M.; Conkrite, K.L.; Sussman, R.T.; Way, G.P.; Adams, R.N.; Sacks, G.I.; Fortina, P.; et al. Epigenomic profiling of neuroblastoma cell lines. Sci. Data 2020, 7, 116. [Google Scholar] [CrossRef] [PubMed]
- Hogarty, M.D.; Norris, M.D.; Davis, K.; Liu, X.; Evageliou, N.F.; Hayes, C.S.; Pawel, B.; Guo, R.; Zhao, H.; Sekyere, E.; et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008, 68, 9735–9745. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Meng, Y.; Zhang, M.; Li, D. MRE11-RAD50-NBS1 complex alterations and DNA damage response: Implications for cancer treatment. Mol. Cancer 2019, 18, 169. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, R.; Tran, S.L.; Maritz, M.F.; Liu, B.; Kong, C.F.; Purgato, S.; Yang, C.; Murray, J.; Russell, A.J.; Flemming, C.L.; et al. MYC-Driven neuroblastomas are addicted to a telomerase-independent function of dyskerin. Cancer Res. 2016, 76, 3604–3617. [Google Scholar] [CrossRef]
- Rihani, A.; Vandesompele, J.; Speleman, F.; Van Maerken, T. Inhibition of CDK4/6 as a novel therapeutic option for neuroblastoma. Cancer Cell Int. 2015, 15, 76. [Google Scholar] [CrossRef]
- Chilamakuri, R.; Rouse, D.C.; Agarwal, S. Inhibition of Polo-like Kinase 1 by HMN-214 blocks cell cycle progression and inhibits neuroblastoma growth. Pharmaceuticals 2022, 15, 523. [Google Scholar] [CrossRef]
- Zimmerman, M.W.; Durbin, A.D.; He, S.; Oppel, F.; Shi, H.; Tao, T.; Li, Z.; Berezovskaya, A.; Liu, Y.; Zhang, J.; et al. Retinoic acid rewiRes. the adrenergic core regulatory circuitry of childhood neuroblastoma. Sci. Adv. 2021, 7, eabe0834. [Google Scholar] [CrossRef]
- Gorlick, R.; Kolb, E.A.; Keir, S.T.; Maris, J.M.; Reynolds, C.P.; Kang, M.H.; Carol, H.; Lock, R.; Billups, C.A.; Kurmasheva, R.T.; et al. Initial testing (stage 1) of the Polo-like kinase inhibitor volasertib (BI 6727), by the Pediatric Preclinical Testing Program. Pediatr. Blood Cancer 2014, 61, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, A.S.; Geerts, D. Polyamine synthesis as a target of MYC oncogenes. J. Biol. Chem 2018, 293, 18757–18769. [Google Scholar] [CrossRef] [PubMed]
- Bello-Fernandez, C.; Packham, G.; Cleveland, J.L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 1993, 90, 7804–7808. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Connolly, T.; Futcher, B.; Beach, D. Human D-type cyclin. Cell 1991, 65, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Maniero, C.; Scudieri, P.; Haris Shaikh, L.; Zhao, W.; Gurnell, M.; Galietta, L.J.V.; Brown, M.J. ANO4 (anoctaMin. 4) is a novel marker of zona glomerulosa that regulates stimulated aldosterone secretion. Hypertension 2019, 74, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Satheesh, N.J.; Busselberg, D. The role of intracellular calcium for the development and treatment of neuroblastoma. Cancers 2015, 7, 823–848. [Google Scholar] [CrossRef]
- Fan, J.; Salathia, N.; Liu, R.; Kaeser, G.E.; Yung, Y.C.; Herman, J.L.; Kaper, F.; Fan, J.B.; Zhang, K.; Chun, J.; et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 2016, 13, 241–244. [Google Scholar] [CrossRef]
- Monclair, T.; Brodeur, G.M.; Ambros, P.F.; Brisse, H.J.; Cecchetto, G.; Holmes, K.; Kaneko, M.; London, W.B.; Matthay, K.K.; Nuchtern, J.G.; et al. The International Neuroblastoma Risk Group (INRG) staging system: An INRG Task Force report. J. Clin. Oncol. 2009, 27, 298–303. [Google Scholar] [CrossRef]
- Carpenter, E.L.; Mosse, Y.P. Targeting ALK in neuroblastoma—Preclinical and clinical advancements. Nat. Rev. Clin. Oncol. 2012, 9, 391–399. [Google Scholar] [CrossRef]
- Voeller, J.; Sondel, P.M. Advances in anti-GD2 immunotherapy for the treatment of high-risk neuroblastoma. J. Pediatr. Hematol. Oncol. 2019, 41, 163–169. [Google Scholar] [CrossRef]
- Applebaum, M.A.; Henderson, T.O.; Lee, S.M.; Pinto, N.; Volchenboum, S.L.; Cohn, S.L. Second malignancies in patients with neuroblastoma: The effects of risk-based therapy. Pediatr. Blood Cancer 2015, 62, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Lucas, J.T., Jr.; McCarville, M.B.; Cooper, D.A.; Doubrovin, M.; Wakefield, D.; Santiago, T.; Li, Y.; Li, X.; Krasin, M.; Santana, V.; et al. Implications of image-defined risk factors and primary-site response on local control and radiation treatment delivery in the management of high-risk neuroblastoma: Is there a role for de-escalation of adjuvant primary-site radiation therapy? Int. J. Radiat. Oncol. Biol. Phys. 2019, 103, 869–877. [Google Scholar] [CrossRef]
- Oberthuer, A.; Berthold, F.; Warnat, P.; Hero, B.; Kahlert, Y.; Spitz, R.; Ernestus, K.; Konig, R.; Haas, S.; Eils, R.; et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J. Clin. Oncol. 2006, 24, 5070–5078. [Google Scholar] [CrossRef] [PubMed]
- Garcia, I.; Mayol, G.; Rios, J.; Domenech, G.; Cheung, N.K.; Oberthuer, A.; Fischer, M.; Maris, J.M.; Brodeur, G.M.; Hero, B.; et al. A three-gene expression signature model for risk stratification of patients with neuroblastoma. Clin. Cancer Res. 2012, 18, 2012–2023. [Google Scholar] [CrossRef] [PubMed]
- Boon, K.; Caron, H.N.; van Asperen, R.; Valentijn, L.; Hermus, M.C.; van Sluis, P.; Roobeek, I.; Weis, I.; Voute, P.A.; Schwab, M.; et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001, 20, 1383–1393. [Google Scholar] [CrossRef]
- Vo, B.T.; Wolf, E.; Kawauchi, D.; Gebhardt, A.; Rehg, J.E.; Finkelstein, D.; Walz, S.; Murphy, B.L.; Youn, Y.H.; Han, Y.G.; et al. The interaction of Myc with Miz1 defines medulloblastoma subgroup identity. Cancer Cell 2016, 29, 5–16. [Google Scholar] [CrossRef]
- Agarwal, P.; Glowacka, A.; Mahmoud, L.; Bazzar, W.; Larsson, L.G.; Alzrigat, M. MYCN amplification is associated with reduced expression of genes encoding the gamma-secretase complex and NOTCH signaling components in neuroblastoma. Int. J. Mol. Sci. 2023, 24, 8141. [Google Scholar] [CrossRef] [PubMed]
- Breit, S.; Schwab, M. Suppression of MYC by high expression of NMYC in human neuroblastoma cells. J. Neurosci. Res. 1989, 24, 21–28. [Google Scholar] [CrossRef]
- Bechmann, N.; Westermann, F.; Eisenhofer, G. HIF and MYC signaling in adrenal neoplasms of the neural crest: Implications for pediatrics. Front. Endocrinol. 2023, 14, 1022192. [Google Scholar] [CrossRef]
- Montemurro, N.; Pahwa, B.; Tayal, A.; Shukla, A.; De Jesus Encarnacion, M.; Ramirez, I.; Nurmukhametov, R.; Chavda, V.; De Carlo, A. Macrophages in Recurrent Glioblastoma as a Prognostic Factor in the Synergistic System of the Tumor Microenvironment. Neurol. Int. 2023, 15, 595–608. [Google Scholar] [CrossRef]
- Zhu, S.; Lee, J.S.; Guo, F.; Shin, J.; Perez-Atayde, A.R.; Kutok, J.L.; Rodig, S.J.; Neuberg, D.S.; Helman, D.; Feng, H.; et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell 2012, 21, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, M.; Patel, A.G.; Dyer, M.A. Neuroblastoma: When differentiation goes awry. Neuron 2022, 110, 2916–2928. [Google Scholar] [CrossRef] [PubMed]
- Soldatov, R.; Kaucka, M.; Kastriti, M.E.; Petersen, J.; Chontorotzea, T.; Englmaier, L.; Akkuratova, N.; Yang, Y.; Haring, M.; Dyachuk, V.; et al. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 2019, 364, eaas9536. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, J.; Yin, J.; Gan, Y.; Xu, S.; Gu, Y.; Huang, W. Alternative approaches to target Myc for cancer treatment. Signal Transduct. Target. Ther. 2021, 6, 117. [Google Scholar] [CrossRef]
- Bassiri, H.; Benavides, A.; Haber, M.; Gilmour, S.K.; Norris, M.D.; Hogarty, M.D. Translational development of difluoromethylornithine (DFMO) for the treatment of neuroblastoma. Transl. Pediatr. 2015, 4, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Monleon, A.; Kryh Oberg, H.; Gaarder, J.; Berbegall, A.P.; Javanmardi, N.; Djos, A.; Ussowicz, M.; Taschner-Mandl, S.; Ambros, I.M.; Ora, I.; et al. Amplification of CDK4 and MDM2: A detailed study of a high-risk neuroblastoma subgroup. Sci. Rep. 2022, 12, 12420. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D.; Sanavia, T.; Jurman, G. Signature literature review reveals AHCY, DPYSL3, and NME1 as the most recurrent prognostic genes for neuroblastoma. BioData Min. 2023, 16, 7. [Google Scholar] [CrossRef]
- De Preter, K.; Vermeulen, J.; Brors, B.; Delattre, O.; Eggert, A.; Fischer, M.; Janoueix-Lerosey, I.; Lavarino, C.; Maris, J.M.; Mora, J.; et al. Accurate outcome prediction in neuroblastoma across independent data sets using a multigene signature. Clin. Cancer Res. 2010, 16, 1532–1541. [Google Scholar] [CrossRef]
- Vermeulen, J.; De Preter, K.; Naranjo, A.; Vercruysse, L.; Van Roy, N.; Hellemans, J.; Swerts, K.; Bravo, S.; Scaruffi, P.; Tonini, G.P.; et al. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: A retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009, 10, 663–671. [Google Scholar] [CrossRef]
- Korber, V.; Stainczyk, S.A.; Kurilov, R.; Henrich, K.O.; Hero, B.; Brors, B.; Westermann, F.; Hofer, T. Neuroblastoma arises in early fetal development and its evolutionary duration predicts outcome. Nat. Genet. 2023, 55, 619–630. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Y.; Alzrigat, M.; Rodriguez-Garcia, A.; Wang, X.; Bexelius, T.S.; Johnsen, J.I.; Arsenian-Henriksson, M.; Liaño-Pons, J.; Bedoya-Reina, O.C. Target Genes of c-MYC and MYCN with Prognostic Power in Neuroblastoma Exhibit Different Expressions during Sympathoadrenal Development. Cancers 2023, 15, 4599. https://doi.org/10.3390/cancers15184599
Yuan Y, Alzrigat M, Rodriguez-Garcia A, Wang X, Bexelius TS, Johnsen JI, Arsenian-Henriksson M, Liaño-Pons J, Bedoya-Reina OC. Target Genes of c-MYC and MYCN with Prognostic Power in Neuroblastoma Exhibit Different Expressions during Sympathoadrenal Development. Cancers. 2023; 15(18):4599. https://doi.org/10.3390/cancers15184599
Chicago/Turabian StyleYuan, Ye, Mohammad Alzrigat, Aida Rodriguez-Garcia, Xueyao Wang, Tomas Sjöberg Bexelius, John Inge Johnsen, Marie Arsenian-Henriksson, Judit Liaño-Pons, and Oscar C. Bedoya-Reina. 2023. "Target Genes of c-MYC and MYCN with Prognostic Power in Neuroblastoma Exhibit Different Expressions during Sympathoadrenal Development" Cancers 15, no. 18: 4599. https://doi.org/10.3390/cancers15184599
APA StyleYuan, Y., Alzrigat, M., Rodriguez-Garcia, A., Wang, X., Bexelius, T. S., Johnsen, J. I., Arsenian-Henriksson, M., Liaño-Pons, J., & Bedoya-Reina, O. C. (2023). Target Genes of c-MYC and MYCN with Prognostic Power in Neuroblastoma Exhibit Different Expressions during Sympathoadrenal Development. Cancers, 15(18), 4599. https://doi.org/10.3390/cancers15184599