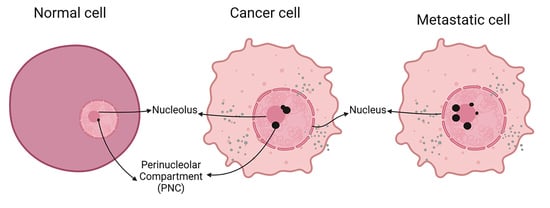
Perinucleolar Compartment (PNC) Prevalence as an Independent Prognostic Factor in Pediatric Ewing Sarcoma: A Multi-Institutional Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Immunohistochemistry
2.3. NanoString miRNA Profiling
2.4. Statistical Analysis
3. Results
3.1. Data for Patient Race and Ethnicity as well as Tumor Metastasis and Recurrence
3.2. Correlation of High PNC Prevalence with Disease Outcome
3.3. Positive and Negative Correlation of miRNAs with High PNC
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riggi, N.; Suvà, M.L.; Stamenkovic, I. Ewing’s Sarcoma. N. Engl. J. Med. 2021, 384, 154–164. [Google Scholar] [CrossRef]
- The WHO Classification of Tumours: Soft Tissue and Bone Tumours, 5th ed.; WHO: Lyon, France, 2020; pp. 323–325. ISBN 978-92-832-4502-5. Available online: https://publications.iarc.fr/588 (accessed on 15 March 2023).
- Mirabello, L.; Troisi, R.J.; Savage, S.A. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int. J. Cancer 2009, 125, 229–234. [Google Scholar] [CrossRef] [Green Version]
- Esiashvili, N.; Goodman, M.; Marcus, R.B. Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: Surveillance Epidemiology and End Results data. J. Pediatr. Hematol. Oncol. 2008, 30, 425–430. [Google Scholar] [CrossRef]
- Isaevska, E.; Manasievska, M.; Alessi, D.; Mosso, M.L.; Magnani, C.; Sacerdote, C.; Pastore, G.; Fagioli, F.; Merletti, F.; Maule, M. Cancer incidence rates and trends among children and adolescents in Piedmont, 1967–2011. PLoS ONE 2017, 12, e0181805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, D.N.; Chastain, K.; Chou, J.F.; Moskowitz, C.S.; Adsuar, R.; Wexler, L.H.; Chou, A.J.; DeRosa, A.; Candela, J.; Magnan, H.; et al. Morbidity and mortality after treatment of Ewing sarcoma: A single-institution experience. Pediatr. Blood Cancer 2017, 64, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Worch, J.; Matthay, K.K.; Neuhaus, J.; Goldsby, R.; Dubois, S.G. Ethnic and racial differences in patients with ewing sarcoma. Cancer 2010, 116, 983–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Mater, D.; Wagner, L. Management of recurrent Ewing sarcoma: Challenges and approaches. Onco Targets Ther. 2019, 12, 2279–2288. [Google Scholar] [CrossRef] [Green Version]
- Slusarczyk, A.; Kamath, R.; Wang, C.; Anchel, D.; Pollock, C.; Lewandowska, M.A.; Fitzpatrick, T.; Bazett-Jones, D.P.; Huang, S. Structure and function of the perinucleolar compartment in cancer cells. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 599–605. [Google Scholar] [CrossRef] [Green Version]
- Norton, J.T.; Wang, C.; Gjidoda, A.; Henry, R.W.; Huang, S. The perinucleolar compartment is directly associated with DNA. J. Biol. Chem. 2009, 284, 4090–4101. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.; Wang, C.; Huang, S. The perinucleolar compartment associates with malignancy. Front. Biol. 2013, 8, 369–376. [Google Scholar] [CrossRef] [Green Version]
- Frankowski, K.J.; Patnaik, S.; Wang, C.; Southall, N.; Dutta, D.; De, S.; Li, D.; Dextras, C.; Lin, Y.-H.; Bryant-Connah, M.; et al. Discovery and Optimization of Pyrrolopyrimidine Derivatives as Selective Disruptors of the Perinucleolar Compartment, a Marker of Tumor Progression toward Metastasis. J. Med. Chem. 2022, 65, 8303–8331. [Google Scholar] [CrossRef]
- Norton, J.T.; Huang, S. The perinucleolar compartment: RNA metabolism and cancer. Cancer Treat. Res. 2013, 158, 139–152. [Google Scholar] [PubMed] [Green Version]
- Huang, S.; Deerinck, T.J.; Ellisman, M.H.; Spector, D.L. The dynamic organization of the perinucleolar compartment in the cell nucleus. J. Cell Biol. 1997, 137, 965–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, S.; Wang, C.; Zhang, C.; Feng, L.; Zhang, W.; Zhou, X.; He, Y.; Xia, X.; Chen, B.; Song, W. PTB: Not just a polypyrimidine tract-binding protein. J. Cell. Physiol. 2022, 237, 2357–2373. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Uchiyama, K.; Akao, Y. PTBP1-targeting microRNAs regulate cancer-specific energy metabolism through the modulation of PKM1/M2 splicing. Cancer Sci. 2021, 112, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Slusarczyk, A.; Huang, S. The perinucleolar Compartment (PNC): Detection by immunohistochemestry. In The Nucleus. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2008; Volume 463. [Google Scholar]
- Kanis, M.J.; Qiang, W.; Pineda, M.; Maniar, K.P.; Kim, J.J. A small molecule inhibitor of the perinucleolar compartment, ML246, attenuates growth and spread of ovarian cancer. Gynecol. Oncol. Res. Pract. 2018, 5, 7. [Google Scholar] [CrossRef]
- Vilimas, T.; Wang, A.Q.; Patnaik, S.; Hughes, E.A.; Singleton, M.D.; Knotts, Z.; Li, D.; Frankowski, K.; Schlomer, J.J.; Guerin, T.M.; et al. Pharmacokinetic evaluation of the PNC disassembler metarrestin in wild-type and Pdx1-Cre;LSL-KrasG12D/+;Tp53R172H/+ (KPC) mice, a genetically engineered model of pancreatic cancer. Cancer Chemother. Pharmacol. 2018, 82, 1067–1080. [Google Scholar] [CrossRef] [Green Version]
- Hennig, C. fpc: Flexible Procedures for Clustering. Bologna. 2020. Available online: https://cran.r-project.org/web/packages/fpc/index.html (accessed on 13 December 2022).
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Koohbanani, B.; Han, G.; Reed, D.; Zhao, Q.; Yi, D. Henderson-Jackson E, Bui MM. Ethnicity and age disparities in Ewing sarcoma outcome. Fetal Pediatr. Pathol. 2013, 32, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Norton, J.T.; Pollock, C.B.; Wang, C.; Schink, J.C.; Kim, J.J.; Huang, S. Perinucleolar compartment prevalence is a phenotypic pancancer marker of malignancy. Cancer 2008, 113, 861–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamath, R.V.; Thor, A.D.; Wang, C.; Edgerton, S.M.; Slusarczyk, A.; Leary, D.J.; Wang, J.; Wiley, E.L.; Jovanovic, B.; Wu, Q.; et al. Perinucleolar compartment prevalence has an independent prognostic value for breast cancer. Cancer Res. 2005, 65, 246–253. [Google Scholar] [CrossRef]
- Frankowski, K.J.; Wang, C.; Patnaik, S.; Schoenen, F.J.; Southall, N.; Li, D.; Teper, Y.; Sun, W.; Kandela, I.; Hu, D.; et al. Metarrestin, a perinucleolar compartment inhibitor, effectively suppresses metastasis. Sci. Transl. Med. 2018, 10, eaap8307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Hoang, B.H.; Ziogas, A.; Zell, J.A. Analysis of prognostic factors in Ewing sarcoma using a population-based cancer registry. Cancer 2010, 116, 1964–1973. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, X.; Shen, J.; Wu, W.K.K.; Chan, M.T.V. MicroRNA expression and its clinical implications in Ewing’s sarcoma. Cell Prolif. 2015, 48, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yufeng, Z.; Ming, Q.; Dandan, W. MiR-320d Inhibits Progression of EGFR-Positive Colorectal Cancer by Targeting TUSC3. Front. Genet. 2021, 12, 738559. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, X.; Pan, B.; He, B.; Chen, X.; Zeng, K.; Xu, M.; Pan, Y.; Sun, H.; Xu, T.; et al. Circulating miR-1290 and miR-320d as Novel Diagnostic Biomarkers of Human Colorectal Cancer. J. Cancer 2019, 10, 43–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]




| Frequencies n = 40 (%) | PNC a Prevalence Median [IQR] | p-Value b | ||
|---|---|---|---|---|
| Sex | Female | 22 (55) | 32.5 [20, 58.75] | 0.763 |
| Male | 18 (45) | 37.5 [20, 81.25] | ||
| Age | <10 years | 13 (32) | 30 [20, 65] | 0.999 |
| ≥10 years | 26 (65) | 35 [20, 77.5] | ||
| Race/Ethnicity | Caucasians | 19 (48) | 25 [20, 40] | 0.212 |
| Hispanics from USA | 6 (15) | 67.5 [46.25, 81.25] | ||
| Mexicans | 14 (35) | 30 [11.25, 90] | ||
| Asians | 1 (2.5) | 85 [85, 85] | ||
| Tumor Volume | n = 31 | 0.459 | ||
| <100 mL | 15 (48) | 30 [20, 50] | ||
| ≥100 mL | 16 (52) | 40 [20, 70] | ||
| Tumor Size | n = 31 | |||
| <8 cm | 13 (42) | 20 [20, 45] | 0.271 | |
| ≥8 cm | 18 (58) | 40 [22.5, 77.5] | ||
| Tumor Necrosis | n = 38 | |||
| <90% | 24 (63) | 30 [20, 65] | 0.643 | |
| ≥90% | 14 (37) | 40 [20, 67.5] | ||
| Metastasis at diagnosis | No | 24 (60) | 20 [18.75, 41.25] | 0.022 |
| Yes | 16 (40) | 62.5 [25, 86.25] | ||
| Progression | n = 38 | |||
| No | 28 (74) | 22.5 [20, 56.25] | 0.079 | |
| Yes | 10 (26) | 62.5 [28.75, 88.75] | ||
| Relapse | n = 38 | |||
| No | 18 (47) | 35 [20, 62.5] | 0.918 | |
| Yes | 20 (53) | 35 [18.75, 70] | ||
| Progression and or Relapse | n = 40 | |||
| No | 12 (30) | 25 [20, 57.5] | 0.656 | |
| Yes | 28 (70) | 37.5 [20, 85] | ||
| Death | n = 40 | |||
| No | 18 (45) | 35 [20, 58.75] | 0.632 | |
| Yes | 22 (55) | 35 [20, 81.25] |
| Hispanics | Caucasians | p Value a | ||
|---|---|---|---|---|
| Diameter | <8 cm | 6 | 7 | 0.710 |
| ≥8 cm | 6 | 12 | ||
| Volume | <100 cm3 | 7 | 8 | 0.473 |
| ≥100 cm3 | 5 | 11 | ||
| Metastasis at diagnosis | No | 12 | 12 | 0.999 |
| Yes | 8 | 7 | ||
| Primary tumor site | No favorable | 11 | 11 | 0.752 |
| Favorable | 9 | 7 | ||
| Tumor necrosis | <90% | 13 | 11 | 0.748 |
| ≥90% | 7 | 8 | ||
| High PNC | No | 12 | 19 | 0.003 |
| Yes | 8 | 0 |
| Model No. 1 | Model No. 2 | |||
|---|---|---|---|---|
| Hazard Ratio | Confidence Limits | Hazard Ratio | Confidence Limits | |
| Metastasis | 3.02 | 1.43, 10.77 | 3.26 | 1.39, 11.51 |
| High PNC | 2.52 | 1.10, 7.74 | ||
| Upregulated miRNA | Downregulated miRNA |
|---|---|
| HSA-miR-490-3p | HSA-miR-1246 |
| HSA-miR-29C-3p | HSA-miR-25-3p |
| HSA-miR-100-5p | HSA-miR-4488 |
| HSA-miR-30A-5p | HSA-miR-320e |
| HSA-miR-30A-3p | HSA-miR-483-5p |
| HSA-miR-152-5p | HSA-miR-320d |
| HSA-miR-3195 | |
| HSA-miR-548ar-5p | |
| HSA-miR-1915-3p | |
| HSA-miR-601 | |
| HSA-miR-1322 | |
| HSA-miR-500a-5p + HSA-miR-501-5p | |
| HSA-miR-892b | |
| HSA-miR-1299 | |
| HSA-miR-1206 | |
| HSA-miR-197-3p | |
| HSA-miR-198 | |
| HSA-miR-660-5p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez, E.; Ahmed, A.A.; McCarthy, L.; Chastain, K.; Habeebu, S.; Zapata-Tarres, M.; Cardenas-Cardos, R.; Velasco-Hidalgo, L.; Corcuera-Delgado, C.; Rodriguez-Jurado, R.; et al. Perinucleolar Compartment (PNC) Prevalence as an Independent Prognostic Factor in Pediatric Ewing Sarcoma: A Multi-Institutional Study. Cancers 2023, 15, 2230. https://doi.org/10.3390/cancers15082230
Gonzalez E, Ahmed AA, McCarthy L, Chastain K, Habeebu S, Zapata-Tarres M, Cardenas-Cardos R, Velasco-Hidalgo L, Corcuera-Delgado C, Rodriguez-Jurado R, et al. Perinucleolar Compartment (PNC) Prevalence as an Independent Prognostic Factor in Pediatric Ewing Sarcoma: A Multi-Institutional Study. Cancers. 2023; 15(8):2230. https://doi.org/10.3390/cancers15082230
Chicago/Turabian StyleGonzalez, Elizabeth, Atif A. Ahmed, Laura McCarthy, Katherine Chastain, Sahibu Habeebu, Marta Zapata-Tarres, Rocio Cardenas-Cardos, Liliana Velasco-Hidalgo, Celso Corcuera-Delgado, Rodolfo Rodriguez-Jurado, and et al. 2023. "Perinucleolar Compartment (PNC) Prevalence as an Independent Prognostic Factor in Pediatric Ewing Sarcoma: A Multi-Institutional Study" Cancers 15, no. 8: 2230. https://doi.org/10.3390/cancers15082230
APA StyleGonzalez, E., Ahmed, A. A., McCarthy, L., Chastain, K., Habeebu, S., Zapata-Tarres, M., Cardenas-Cardos, R., Velasco-Hidalgo, L., Corcuera-Delgado, C., Rodriguez-Jurado, R., García-Rodríguez, L., Parrales, A., Iwakuma, T., Farooqi, M. S., Lee, B., Weir, S. J., & Flatt, T. G. (2023). Perinucleolar Compartment (PNC) Prevalence as an Independent Prognostic Factor in Pediatric Ewing Sarcoma: A Multi-Institutional Study. Cancers, 15(8), 2230. https://doi.org/10.3390/cancers15082230