The miR-141/200c-STAT4 Axis Contributes to Leukemogenesis by Enhancing Cell Proliferation in T-PLL
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. Cell Culture
2.3. Stable miR-141/200c-Overexpressing Cell Lines
2.4. RNA Isolation and Sequencing, Data Processing and Analyses, and miR Target Prediction
2.5. Western Blot and Quantitative Real-Time PCR
2.6. Assessment of Associations with Surface Marker Expression and Clinical Data
3. Results
3.1. The Micro-RNA Cluster miR-141/200c Is Highly Upregulated in T-PLL Cells and Is Associated with a Gene Signature of Cell Cycle Acceleration and Enhanced Survival Signaling
3.2. Elevated Expression of miR-141/200c Leads to Enhanced Cell Proliferation and Reduced Stress-Induced Cell Death under Serum Starvation in Mature T-Cell Leukemia/Lymphoma Lines
3.3. Overexpression of miR-141/200c Shapes a Pro-Oncogenic Transcriptome of the Mature T-Cell Lymphoma Line HuT78, Resembling the Alterations Seen in T-PLL
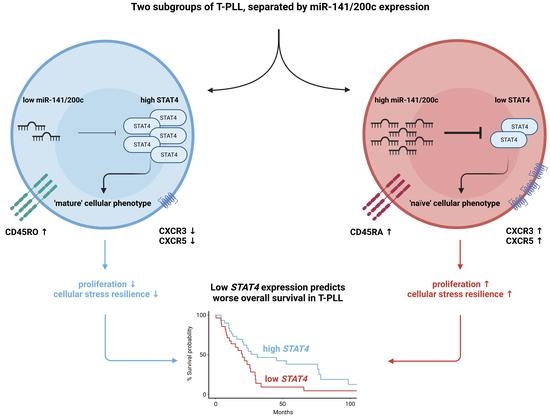
3.4. Downregulation of STAT4 by the miR-141/200c Cluster Is Associated with an Immature Cell Phenotype and Poor Patient Outcomes in T-PLL
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jain, P.; Aoki, E.; Keating, M.; Wierda, W.G.; O’Brien, S.; Gonzalez, G.N.; Ferrajoli, A.; Jain, N.; Thompson, P.A.; Jabbour, E.; et al. Characteristics, Outcomes, Prognostic Factors and Treatment of Patients with T-Cell Prolymphocytic Leukemia (T-PLL). Ann. Oncol. 2017, 28, 1554–1559. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.d.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Staber, P.B.; Herling, M.; Bellido, M.; Jacobsen, E.D.; Davids, M.S.; Kadia, T.M.; Shustov, A.; Tournilhac, O.; Bachy, E.; Zaja, F.; et al. Consensus Criteria for Diagnosis, Staging, and Treatment Response Assessment of T-Cell Prolymphocytic Leukemia. Blood 2019, 134, 1132–1143. [Google Scholar] [CrossRef]
- Braun, T.; von Jan, J.; Wahnschaffe, L.; Herling, M. Advances and Perspectives in the Treatment of T-PLL. Curr. Hematol. Malig. Rep. 2020, 15, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Herling, M.; Patel, K.A.; Teitell, M.A.; Konopleva, M.; Ravandi, F.; Kobayashi, R.; Jones, D. High TCL1 Expression and Intact T-Cell Receptor Signaling Define a Hyperproliferative Subset of T-Cell Prolymphocytic Leukemia. Blood 2008, 111, 328–337. [Google Scholar] [CrossRef]
- Schrader, A.; Crispatzu, G.; Oberbeck, S.; Mayer, P.; Pützer, S.; von Jan, J.; Vasyutina, E.; Warner, K.; Weit, N.; Pflug, N.; et al. Actionable Perturbations of Damage Responses by TCL1/ATM and Epigenetic Lesions Form the Basis of T-PLL. Nat. Commun. 2018, 9, 697. [Google Scholar] [CrossRef] [PubMed]
- Wahnschaffe, L.; Braun, T.; Timonen, S.; Giri, A.K.; Schrader, A.; Wagle, P.; Almusa, H.; Johansson, P.; Bellanger, D.; López, C.; et al. JAK/STAT-Activating Genomic Alterations Are a Hallmark of T-PLL. Cancers 2019, 11, 1833. [Google Scholar] [CrossRef] [PubMed]
- Nishikomori, R.; Usui, T.; Wu, C.-Y.; Morinobu, A.; O’Shea, J.J.; Strober, W. Activated STAT4 Has an Essential Role in Th1 Differentiation and Proliferation That Is Independent of Its Role in the Maintenance of IL-12Rβ2 Chain Expression and Signaling. J. Immunol. 2002, 169, 4388–4398. [Google Scholar] [CrossRef]
- Kim, K.; Lee, Y.S.; Carthew, R.W. Conversion of Pre-RISC to Holo-RISC by Ago2 during Assembly of RNAi Complexes. RNA 2007, 13, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Stachelscheid, J.; Bley, N.; Oberbeck, S.; Otte, M.; Müller, T.A.; Wahnschaffe, L.; Glaß, M.; Ommer, K.; Franitza, M.; et al. Noncanonical Function of AGO2 Augments T-Cell Receptor Signaling in T-Cell Prolymphocytic Leukemia. Cancer Res. 2022, 82, 1818–1831. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 Mediates RNA Cleavage Targeted by MiRNAs and SiRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. MiR-15 and MiR-16 Induce Apoptosis by Targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.J.; Factora, T.D.; Dey, S.; Kota, J. A Systematic Review of MiR-29 in Cancer. Mol. Ther. Oncolytics 2018, 12, 173–194. [Google Scholar] [CrossRef]
- Shen, X.; Wang, B.; Li, K.; Wang, L.; Zhao, X.; Xue, F.; Shi, R.; Zheng, J. MicroRNA Signatures in Diagnosis and Prognosis of Cutaneous T Cell Lymphoma. J. Investig. Dermatol. 2018, 138, 202. [Google Scholar] [CrossRef]
- Wu, S.J.; Chen, J.; Wu, B.; Wang, Y.J.; Guo, K.Y. MicroRNA-150 Enhances Radiosensitivity by Inhibiting the AKT Pathway in NK/T Cell Lymphoma. J. Exp. Clin. Cancer Res. 2018, 37, 18. [Google Scholar] [CrossRef]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating Cancer with MicroRNA Replacement Therapy: A Literature Review. J. Cell. Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef]
- Braun, T.; Glass, M.; Wahnschaffe, L.; Otte, M.; Mayer, P.; Franitza, M.; Altmüller, J.; Hallek, M.; Hüttelmaier, S.; Schrader, A.; et al. Micro-RNA Networks in T-Cell Prolymphocytic Leukemia Reflect T-Cell Activation and Shape DNA Damage Response and Survival Pathways. Haematologica 2022, 107, 187. [Google Scholar] [CrossRef]
- Cavallari, I.; Ciccarese, F.; Sharova, E.; Urso, L.; Raimondi, V.; Silic-Benussi, M.; D’Agostino, D.M.; Ciminale, V. The MiR-200 Family of MicroRNAs: Fine Tuners of Epithelial-Mesenchymal Transition and Circulating Cancer Biomarkers. Cancers 2021, 13, 5874. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The MiR-200 Family and MiR-205 Regulate Epithelial to Mesenchymal Transition by Targeting ZEB1 and SIP1. Nat. Cell. Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Yang, C. The MicroRNA-200 Family: Small Molecules with Novel Roles in Cancer Development, Progression and Therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef] [PubMed]
- Erkeland, S.J.; Stavast, C.J.; Schilperoord-Vermeulen, J.; Dal Collo, G.; van de Werken, H.J.G.; Leon, L.G.; van Hoven-Beijen, A.; van Zuijen, I.; Mueller, Y.M.; Bindels, E.M.; et al. The MiR-200c/141-ZEB2-TGFβ Axis Is Aberrant in Human T-Cell Prolymphocytic Leukemia. Haematologica 2022, 107, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Herling, M.; Khoury, J.D.; Washington, L.T.; Duvic, M.; Keating, M.J.; Jones, D. A Systematic Approach to Diagnosis of Mature T-Cell Leukemias Reveals Heterogeneity among WHO Categories. Blood 2004, 104, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Heavican, T.B.; Bouska, A.; Yu, J.; Lone, W.; Amador, C.; Gong, Q.; Zhang, W.; Li, Y.; Dave, B.J.; Nairismägi, M.-L.; et al. Genetic Drivers of Oncogenic Pathways in Molecular Subgroups of Peripheral T-Cell Lymphoma. Blood 2019, 133, 1664–1676. [Google Scholar] [CrossRef]
- Iqbal, J.; Wright, G.; Wang, C.; Rosenwald, A.; Gascoyne, R.D.; Weisenburger, D.D.; Greiner, T.C.; Smith, L.; Guo, S.; Wilcox, R.A.; et al. Gene Expression Signatures Delineate Biological and Prognostic Subgroups in Peripheral T-Cell Lymphoma. Blood 2014, 123, 2915–2923. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Cazes, A.; Childers, B.G.; Esparza, E.; Lowy, A.M. The MST1R/RON Tyrosine Kinase in Cancer: Oncogenic Functions and Therapeutic Strategies. Cancers 2022, 14, 2037. [Google Scholar] [CrossRef]
- Qiuping, Z.; Jei, X.; Youxin, J.; Wei, J.; Chun, L.; Jin, W.; Qun, W.; Yan, L.; Chunsong, H.; Mingzhen, Y.; et al. CC Chemokine Ligand 25 Enhances Resistance to Apoptosis in CD4+ T Cells from Patients with T-Cell Lineage Acute and Chronic Lymphocytic Leukemia by Means of Livin Activation. Cancer Res. 2004, 64, 7579–7587. [Google Scholar] [CrossRef]
- Mohanty, A.; Sandoval, N.; Das, M.; Pillai, R.; Chen, L.; Chen, R.W.; Amin, H.M.; Wang, M.; Marcucci, G.; Weisenburger, D.D.; et al. CCND1 Mutations Increase Protein Stability and Promote Ibrutinib Resistance in Mantle Cell Lymphoma. Oncotarget 2016, 7, 73558–73572. [Google Scholar] [CrossRef] [PubMed]
- Barclay, J.L.; Anderson, S.T.; Waters, M.J.; Curlewis, J.D. SOCS3 as a Tumor Suppressor in Breast Cancer Cells, and Its Regulation by PRL. Int. J. Cancer 2009, 124, 1756–1766. [Google Scholar] [CrossRef] [PubMed]
- Wurster, A.L.; Tanaka, T.; Grusby, M.J. The Biology of Stat4 and Stat6. Oncogene 2000, 19, 2577–2584. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Nakano, K.; Watanabe, E.; Ishigaki, T.; Ohno, N.; Yuji, K.; Oyaizu, N.; Asanuma, S.; Yamagishi, M.; Yamochi, T.; et al. CADM1 Expression and Stepwise Downregulation of CD7 Are Closely Associated with Clonal Expansion of HTLV-I-Infected Cells in Adult T-Cell Leukemia/Lymphoma. Clin. Cancer Res. 2014, 20, 2851–2861. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Silvestris, N.; Mohammadi, A.; Khaze, V.; Baghbani, E.; Mokhtarzadeh, A.; Shanehbandi, D.; Derakhshani, A.; Duijf, P.H.G.; Baradaran, B. MiR-34a and MiR-200c Have an Additive Tumor-Suppressive Effect on Breast Cancer Cells and Patient Prognosis. Genes 2021, 12, 267. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, S.-T.; Wang, X.; Deng, J.; Li, W.-H.; Zhang, P.; Liu, B.-S. MiR-200c Regulates Tumor Growth and Chemosensitivity to Cisplatin in Osteosarcoma by Targeting AKT2. Sci. Rep. 2017, 7, 13598. [Google Scholar] [CrossRef]
- Carter, J.V.; O’Brien, S.J.; Burton, J.F.; Oxford, B.G.; Stephen, V.; Hallion, J.; Bishop, C.; Galbraith, N.J.; Eichenberger, M.R.; Sarojini, H.; et al. The MicroRNA-200 Family Acts as an Oncogene in Colorectal Cancer by Inhibiting the Tumor Suppressor RASSF2. Oncol. Lett. 2019, 18, 3994–4007. [Google Scholar] [CrossRef]
- Huang, G.-L.; Sun, J.; Lu, Y.; Liu, Y.; Cao, H.; Zhang, H.; Calin, G.A. MiR-200 Family and Cancer: From a Meta-Analysis View. Mol. Aspects Med. 2019, 70, 57–71. [Google Scholar] [CrossRef]
- Mao, Y.; Chen, W.; Wu, H.; Liu, C.; Zhang, J.; Chen, S. Mechanisms and Functions of MiR-200 Family in Hepatocellular Carcinoma. Onco Targets Ther. 2020, 13, 13479–13490. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Lahesmaa, R.; Vahedi, G.; Laurence, A.; Kanno, Y. Genomic Views of STAT Function in CD4+ T Helper Cell Differentiation. Nat. Rev. Immunol. 2011, 11, 239–250. [Google Scholar] [CrossRef]
- Cattaneo, F.; Patrussi, L.; Capitani, N.; Frezzato, F.; D’Elios, M.M.; Trentin, L.; Semenzato, G.; Baldari, C.T. Expression of the P66Shc Protein Adaptor Is Regulated by the Activator of Transcription STAT4 in Normal and Chronic Lymphocytic Leukemia B Cells. Oncotarget 2016, 7, 57086–57098. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, T.R.; Nishizawa, Y. Stat4 Suppresses the Proliferation of Connective Tissue-Type Mast Cells. Lab. Investig. 2008, 88, 856–864. [Google Scholar] [CrossRef]
- Wang, G. Decreased STAT4 Indicates Poor Prognosis and Enhanced Cell Proliferation in Hepatocellular Carcinoma. World J. Gastroenterol. 2015, 21, 3983. [Google Scholar] [CrossRef] [PubMed]
- Ebersbach, C.; Beier, A.-M.K.; Thomas, C.; Erb, H.H.H. Impact of STAT Proteins in Tumor Progress and Therapy Resistance in Advanced and Metastasized Prostate Cancer. Cancers 2021, 13, 4854. [Google Scholar] [CrossRef]
- Litvinov, I.V.; Cordeiro, B.; Fredholm, S.; Ødum, N.; Zargham, H.; Huang, Y.; Zhou, Y.; Pehr, K.; Kupper, T.S.; Woetmann, A.; et al. Analysis of STAT4 Expression in Cutaneous T-Cell Lymphoma (CTCL) Patients and Patient-Derived Cell Lines. Cell Cycle 2014, 13, 2975–2982. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Shao, H.; Chen, H.; Deng, Q. The Mechanism of MiR-141 Regulating the Proliferation and Metastasis of Liver Cancer Cells by Targeting STAT4. J. Oncol. 2021, 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, J.; Chen, Q.; Cheng, J.; Chen, X.; Mao, Y.; Chen, W.; Liu, C.; Wu, H.; Lv, Y.; et al. MicroRNA-200a and MicroRNA-141 Have a Synergetic Effect on the Suppression of Epithelial-mesenchymal Transition in Liver Cancer by Targeting STAT4. Oncol. Lett. 2020, 21, 137. [Google Scholar] [CrossRef] [PubMed]
- Lanford, R.E.; Hildebrandt-Eriksen, E.S.; Petri, A.; Persson, R.; Lindow, M.; Munk, M.E.; Kauppinen, S.; Ørum, H. Therapeutic Silencing of MicroRNA-122 in Primates with Chronic Hepatitis C Virus Infection. Science 2010, 327, 198–201. [Google Scholar] [CrossRef]
- Christopher, A.F.; Kaur, R.P.; Kaur, G.; Kaur, A.; Gupta, V.; Bansal, P. MicroRNA Therapeutics: Discovering Novel Targets and Developing Specific Therapy. Perspect. Clin. Res. 2016, 7, 68–74. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The MultiMiR R Package and Database: Integration of MicroRNA–Target Interactions along with Their Disease and Drug Associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otte, M.; Stachelscheid, J.; Glaß, M.; Wahnschaffe, L.; Jiang, Q.; Lone, W.; Ianevski, A.; Aittokallio, T.; Iqbal, J.; Hallek, M.; et al. The miR-141/200c-STAT4 Axis Contributes to Leukemogenesis by Enhancing Cell Proliferation in T-PLL. Cancers 2023, 15, 2527. https://doi.org/10.3390/cancers15092527
Otte M, Stachelscheid J, Glaß M, Wahnschaffe L, Jiang Q, Lone W, Ianevski A, Aittokallio T, Iqbal J, Hallek M, et al. The miR-141/200c-STAT4 Axis Contributes to Leukemogenesis by Enhancing Cell Proliferation in T-PLL. Cancers. 2023; 15(9):2527. https://doi.org/10.3390/cancers15092527
Chicago/Turabian StyleOtte, Moritz, Johanna Stachelscheid, Markus Glaß, Linus Wahnschaffe, Qu Jiang, Waseem Lone, Aleksandr Ianevski, Tero Aittokallio, Javeed Iqbal, Michael Hallek, and et al. 2023. "The miR-141/200c-STAT4 Axis Contributes to Leukemogenesis by Enhancing Cell Proliferation in T-PLL" Cancers 15, no. 9: 2527. https://doi.org/10.3390/cancers15092527
APA StyleOtte, M., Stachelscheid, J., Glaß, M., Wahnschaffe, L., Jiang, Q., Lone, W., Ianevski, A., Aittokallio, T., Iqbal, J., Hallek, M., Hüttelmaier, S., Schrader, A., Braun, T., & Herling, M. (2023). The miR-141/200c-STAT4 Axis Contributes to Leukemogenesis by Enhancing Cell Proliferation in T-PLL. Cancers, 15(9), 2527. https://doi.org/10.3390/cancers15092527