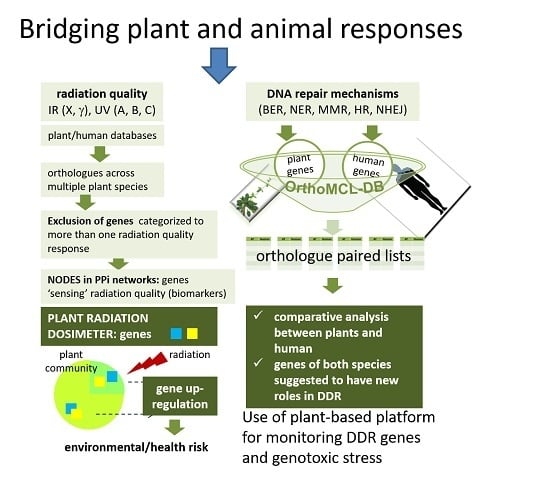
Bridging Plant and Human Radiation Response and DNA Repair through an In Silico Approach
Abstract
:1. Introduction
2. Plant Radiation Biodosimeter
2.1. Bioinformatics Approaches for the Identification of Candidate Genes for the Plant Radiation Dosimeter
2.2. Screening Strategy and Final Selection of Candidate Genes
2.2.1. Selection of Gene Ontology Terms
2.2.2. Orthologous Genes
2.2.3. Exclusion of Common Genes
2.2.4. Protein-Protein Interactions Network
2.2.5. Ranking Based on Multiplicity
2.2.6. Human Orthologues of the Resulting Genes
3. Comparison between Arabidopsis thaliana and Homo sapiens DNA Repair Mechanisms
3.1. Comparative Analysis Strategy
3.1.1. Selection of Gene Ontology Terms
3.1.2. Human DNA Repair Genes
3.1.3. Arabidopsis thaliana DNA Repair Genes
3.1.4. Identification of Orthologies between Homo sapiens and Arabidopsis thaliana DNA Repair Genes
3.2. ‘New Genes’ Emerging from Comparative Analysis
4. An In Vitro Approach Monitoring Key DNA Repair Genes
5. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
Abbreviations
| DDR | DNA damage response |
| SSB | single-strand breaks |
| DSB | double-strand breaks |
| NER | nucleotide excision repair |
| BER | base excision repair |
| MMR | Mismatch repair |
| HR | homologous recombination |
| NHEJ | non-homologous end joining |
| UV | ultraviolet radiation |
| IR | ionizing radiation |
| PAXX | PAralog of XRCC4 and XLF |
| REM | radiation exposure monitoring |
References
- Elledge, S.J. The DNA damage response--Self-awareness for DNA: The 2015 albert lasker basic medical research award. JAMA 2015, 314, 1111–1112. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, P.; Jonkers, J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer 2012, 12, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Jeggo, P.A.; Pearl, L.H.; Carr, A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 2016, 16, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, A.G.; O’Neill, P.; Stewart, R.D. Induction and repair of clustered DNA lesions: What do we know so far? Radiat. Res. 2013, 180, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Ochi, T.; Blackford, A.N.; Coates, J.; Jhujh, S.; Mehmood, S.; Tamura, N.; Travers, J.; Wu, Q.; Draviam, V.M.; Robinson, C.V.; et al. DNA repair. Paxx, a paralog of xrcc4 and xlf, interacts with ku to promote DNA double-strand break repair. Science 2015, 347, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Raschella, G.; Melino, G.; Malewicz, M. New factors in mammalian DNA repair-the chromatin connection. Oncogene 2017. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Jackson, S.P. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011, 25, 409–433. [Google Scholar] [CrossRef] [PubMed]
- Zeegers, D.; Venkatesan, S.; Koh, S.W.; Low, G.K.; Srivastava, P.; Sundaram, N.; Sethu, S.; Banerjee, B.; Jayapal, M.; Belyakov, O.; et al. Biomarkers of ionizing radiation exposure: A multiparametric approach. Genome Integr. 2017, 8, 6. [Google Scholar] [PubMed]
- Shiloh, Y.; Ziv, Y. The atm protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013, 14, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Culligan, K.M.; Robertson, C.E.; Foreman, J.; Doerner, P.; Britt, A.B. Atr and atm play both distinct and additive roles in response to ionizing radiation. Plant J. Cell Mol. Biol. 2006, 48, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Borras-Fresneda, M.; Barquinero, J.F.; Gomolka, M.; Hornhardt, S.; Rossler, U.; Armengol, G.; Barrios, L. Differences in DNA repair capacity, cell death and transcriptional response after irradiation between a radiosensitive and a radioresistant cell line. Sci. Rep. 2016, 6, 27043. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.Y.; Kojic, M.; Holloman, W.K.; Lue, N.F. Brh2 and rad51 promote telomere maintenance in ustilago maydis, a new model system of DNA repair proteins at telomeres. DNA Repair 2013, 12, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.W.; Yang, D.H.; Kim, M.K.; Seo, H.S.; Lim, S.; Bahn, Y.S. Unraveling fungal radiation resistance regulatory networks through the genome-wide transcriptome and genetic analyses of cryptococcus neoformans. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Pfister, T.D.; Parchment, R.E.; Kummar, S.; Rubinstein, L.; Evrard, Y.A.; Gutierrez, M.E.; Murgo, A.J.; Tomaszewski, J.E.; Doroshow, J.H.; et al. Monitoring drug-induced gammah2ax as a pharmacodynamic biomarker in individual circulating tumor cells. Clin. Cancer Res. 2010, 16, 1073–1084. [Google Scholar] [CrossRef] [PubMed]
- Schoenfelder, K.P.; Fox, D.T. The expanding implications of polyploidy. J. Cell Biol. 2015, 209, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Geras’kin, S.A. Ecological effects of exposure to enhanced levels of ionizing radiation. J. Environ. Radioact. 2016, 162–163, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Bin, P.; Zheng, Y. Association between gamma-h2ax and DNA double-strand breaks. Wei Sheng Yan Jiu 2007, 36, 520–522. [Google Scholar] [PubMed]
- Nikitaki, Z.; Hellweg, C.; Georgakilas, A.G.; Ravanat, J.L. Stress-induced DNA damage biomarkers: Applications and limitations. Front. Chem. 2015, 3, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Nikitaki, Z.; Mavragani, I.V.; Laskaratou, D.A.; Gika, V.; Moskvin, V.P.; Theofilatos, K.; Vougas, K.; Stewart, R.D.; Georgakilas, A.G. Systemic mechanisms and effects of ionizing radiation: A new ‘old’ paradigm of how the bystanders and distant can become the players. Semin. Cancer Biol. 2016, 37–38, 77–95. [Google Scholar] [CrossRef] [PubMed]
- Pateras, I.S.; Havaki, S.; Nikitopoulou, X.; Vougas, K.; Townsend, P.A.; Panayiotidis, M.I.; Georgakilas, A.G.; Gorgoulis, V.G. The DNA damage response and immune signaling alliance: Is it good or bad? Nature decides when and where. Pharmacol. Ther. 2015, 154, 36–56. [Google Scholar] [CrossRef] [PubMed]
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O'Donovan, C.; Apweiler, R. Quickgo: A web-based tool for gene ontology searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef] [PubMed]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 2016, 1374, 115–140. [Google Scholar] [PubMed]
- Bioinformatics & Evolutionary Genomics. Calculate and Draw Custom Venn Diagrams. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 6 October 2016).
- Lobachevsky, P.N.; Martin, R.F. Plasmid DNA breakage by decay of DNA-associated auger emitters: Experiments with 123i/125i-iodohoechst 33258. Int. J. Radiat. Biol. 2004, 80, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. Ncbi reference sequence (refseq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005, 33, D501–D504. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.; Barker, D.; Birney, E.; Cameron, G.; Chen, Y.; Clark, L.; Cox, T.; Cuff, J.; Curwen, V.; Down, T.; et al. The ensembl genome database project. Nucleic Acids Res. 2002, 30, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Povey, S.; Lovering, R.; Bruford, E.; Wright, M.; Lush, M.; Wain, H. The hugo gene nomenclature committee (hgnc). Hum. Genet. 2001, 109, 678–680. [Google Scholar] [CrossRef] [PubMed]
- Lobachevsky, P.N.; White, J.; Leung, M.; Skene, C.; White, J.; Martin, R.F. Plasmid breakage by (125)i-labelled DNA ligands: Effect of DNA-iodine atom distance on breakage efficiency. Int. J. Radiat. Biol. 2008, 84, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Mackey, A.J.; Stoeckert, C.J., Jr.; Roos, D.S. Orthomcl-db: Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006, 34, D363–D368. [Google Scholar] [CrossRef] [PubMed]
- Iliakis, G.; Murmann, T.; Soni, A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 793, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Welcsh, P.L.; King, M.C. Brca1 and brca2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet 2001, 10, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yu, X. Function of brca1 in the DNA damage response is mediated by adp-ribosylation. Cancer Cell 2013, 23, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Dantzer, F.; Ame, J.C.; Schreiber, V.; Nakamura, J.; Menissier-de Murcia, J.; de Murcia, G. Poly(adp-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006, 409, 493–510. [Google Scholar] [PubMed]
- Angelis, K.J.; Dusinska, M.; Collins, A.R. Single cell gel electrophoresis: Detection of DNA damage at different levels of sensitivity. Electrophoresis 1999, 20, 2133–2138. [Google Scholar] [CrossRef]
- Hedges, S.B.; Dudley, J.; Kumar, S. Timetree: A public knowledge-base of divergence times among organisms. Bioinformatics 2006, 22, 2971–2972. [Google Scholar] [CrossRef] [PubMed]
- Stover, E.H.; Konstantinopoulos, P.A.; Matulonis, U.A.; Swisher, E.M. Biomarkers of response and resistance to DNA repair targeted therapies. Clin. Cancer Res. 2016, 22, 5651–5660. [Google Scholar] [CrossRef] [PubMed]












| GO Term | Annotation | Category |
|---|---|---|
| GO:0010212 | response to ionizing radiation | Ionizing Radiation |
| GO:0010165 | response to X-ray | X-ray |
| GO:0010332 | response to gamma radiation | γ-ray |
| GO:0009411 | response to UV | UV |
| GO:0070141 | response to UV-A | UV-A |
| GO:0010224 | response to UV-B | UV-B |
| GO:0010225 | response to UV-C | UV-C |
| γ-rays | # | X-rays | # | UV-A | # | UV-B | # | UV-C | # |
|---|---|---|---|---|---|---|---|---|---|
| RAD54 | 30 | ATLIG4 | 28 | AT2G21970.1 | 2 | UVH3 | 32 | TED4 | 3 |
| AT4G14970 | 30 | MIM | 7 | RPA70B | 7 | MC4 | 2 | ||
| RPA1A | 27 | SMC6A | 6 | RPA70D | 5 | MC7 | 2 | ||
| MSH5 | 23 | XPB1 | 4 | MC5 | 2 | ||||
| RAD51 | 18 | XPB2 | 4 | MC6 | 2 | ||||
| AT3G48900 | 1 | MC8 | 2 | ||||||
| HO4 | 1 | ||||||||
| HO3 | 1 | ||||||||
| ETL1 | 1 | ||||||||
| PCC1 | 1 | ||||||||
| PAD4 | 1 |
| Type of Radiation | Arabidopsis thaliana | Ortho Group | Homo Sapiens | ||||
|---|---|---|---|---|---|---|---|
| TAIR | Gene Name | RefSeq | ENSP | HGNC | GO | ||
| γ-rays | RAD54 | AT3G19210 | NP_188552 | OG5_127098 | ENSP00000336606 | RAD54B | GO:0010212 |
| ENSP00000396113 | RAD54L | GO:0010212 | |||||
| AT4G14970 | NP_193233 | OG5_132711 | ENSP00000287647 | FANCD2 | GO:0010332 | ||
| RPA1A | AT2G06510 | NP_973433 | OG5_127539 | ENSP00000254719 | RPA1 | -- | |
| MSH5 | AT3G20475 | NP_188683 | OG5_129379 | ENSP00000364894 | MSH5 | -- | |
| ENSP00000387668 | MSH5 | ||||||
| ENSP00000394619 | MSH5 | ||||||
| ENSP00000394649 | MSH5 | ||||||
| ENSP00000406868 | MSH5 | ||||||
| ENSP00000407047 | MSH5 | ||||||
| ENSP00000409207 | MSH5 | ||||||
| RAD51 | AT1G07745 | NP_172254 | OG5_132909 | ENSP00000378090 | RAD51D | GO:0010212 | |
| X-rays | ATLIG4 | AT5G57160 | NP_568851 | OG5_130132 | ENSP00000402030 | LIG4 | GO:0010212 GO:0010165 GO:0010332 |
| MIM | AT5G61460 | NP_200954 | OG5_127751 | ENSP00000370672 | SMC6 | GO:0010165 | |
| SMC6A | AT5G07660 | NP_196383 | OG5_127751 | ENSP00000370672 | SMC6 | -- | |
| UV-A | SEP2 | AT2G21970 | NP_565524 | OG5_178242 | no | -- | -- |
| UV-B | UVH3 | AT3G28030 | NP_566830 | OG5_128675 | ENSP00000347978 | ERCC5 | GO:0009411 GO:0010225 |
| RPA70B | AT5G08020 | NP_196419 | OG5_127539 | ENSP00000254719 | RPA1 | -- | |
| RPA70D | AT5G61000 | NP_200908 | OG5_127539 | ENSP00000254719 | RPA1 | -- | |
| XPB2 | AT5G41360 | NP_568591 | OG5_127208 | ENSP00000285398 | ERCC3 | GO:0009411 | |
| XPB1 | AT5G41370 | NP_568592 | OG5_127208 | ENSP00000285398 | ERCC3 | -- | |
| GEN2 | AT3G48900 | NP_001118795 | OG5_174560 | no | -- | -- | |
| UV-C | TED4 | AT2G26670 | NP_001118392 | OG5_140322 | no | -- | -- |
| MC4 | AT1G79340 | NP_178052 | OG5_147205 | no | -- | -- | |
| MC8 | AT1G16420 | NP_173092 | OG5_134790 | no | -- | -- | |
| MC7 | AT1G79310 | NP_178049 | 0 | -- | -- | ||
| MC5 | AT1G79330 | NP_178051 | OG5_147205 | no | -- | -- | |
| MC6 | AT1G79320 | NP_178050 | 0 | -- | -- | ||
| HO4 | AT1G58300 | NP_176126 | OG5_140322 | no | -- | -- | |
| HO3 | AT1G69720 | NP_177130 | OG5_140322 | no | -- | -- | |
| ETL1 | AT2G02090 | NP_178318 | OG5_129286 | ENSP00000351947 | SMARCAD1 | -- | |
| PCC1 | AT3G22231 | NP_566702 | OG5_144902 | no | -- | -- | |
| PAD4 | AT3G52430 | NP_190811 | OG5_190312 | no | -- | -- | |
| GO Term | Annotation | Abbreviation |
|---|---|---|
| GO:0006281 | DNA repair | DNA repair |
| GO:0006284 | base-excision repair | BER |
| GO:0006289 | nucleotide-excision repair | NER |
| GO:0006298 | mismatch repair | MMR |
| GO:0000724 | double-strand break repair via homologous recombination | HR |
| GO:0006303 | double-strand break repair via non-homologous end joining | NHEJ |
| Homo sapiens | Arabidopsis thaliana | Homo sapiens | Arabidopsis thaliana | Homo sapiens | Arabidopsis thaliana | Homo sapiens | Arabidopsis thaliana |
|---|---|---|---|---|---|---|---|
| ERCC6 | CHR8 | GINS4 | SLD5 | GTF2H4 | AT4G17020 | ALKBH3 | ALKBH2 |
| NSMCE2 | MMS21 | UNG | ATUNG | DCLRE1A | SNM1 | XRCC1 | XRCC1 |
| RNASEH2A | AT2G25100 | APTX | BHLH140 | XRCC5 | KU80 | XRCC2 | XRCC2 |
| TOP3A | TOP3A | ALKBH1 | AT1G11780 | NSMCE1 | emb1379 | XRCC4 | XRCC4 |
| RECQL RECQL5 WRN BLM | RECQL4A MED34 | RAD23A RAD23B | RAD23A RAD23B RAD23C RAD23D | RPA1 | RPA1A RPA1B RPA1C RPA1D | ERCC5 | UVH3 |
| OGG1 | OGG1 | ||||||
| ERCC6L2 | SWI2 | ||||||
| ERCC1 | ERCC1 | ||||||
| KAT5 | HAM2 HAM1 | DDB1 | DDB1A DDB1B | KIF22 | AT5G02370 | ASF1A | ASF1A ASF1B |
| MRE11A | MRE11 | ||||||
| RAD51C | RAD51C | XRCC3 | XRCC3 | DDB2 | DDB2 | GEN1 | GEN1 |
| EXO1 | AT1G18090 | MPG | MAG | RAD1 | AT4G17760 | REV3L | REV3 |
| APEX2 | ARP APE1L APE2 | UBE2V1 | UEV1A UEV1B UEV1C | RPS3 | RPS3A RPS3B RPS3C | UBE2A UBE2B | UBC1 UBC2 UBC3 |
| MLH1 | MLH1 | SHFM1 | ATDSS1(V) | POLD3 | POLD3 | XPC | ATRAD4 |
| MTOR | TOR | SSRP1 | SSRP1 | XRCC6 | KU70 | ZSWIM7 | AT4G33925 |
| CUL4ACUL4B | CUL4 | DMAP1 | SWC4 | TDP1 | TDP1 | ERCC4 | UVH1 |
| MCM8 | MCM8 | NSMCE4A | NSE4A | ERCC2 | UVH6 | ||
| RRM2B | RNR2A TSO2 | SLX1A SLX1B | AT2G30350 | DMC1 RAD51 | DMC1 RAD51 | ERCC3 | XPB2 XPB1 |
| CDC5L | CDC5 | ACTL6A | ARP4 | PARP2 | PARP2 | BRCA2 | BRCA2B |
| PMS2 | PMS1 | PNKP | ZDP | GINS2 | GINS2 | POLL | AT1G10520 |
| SUPT16H | SPT16 | ATR | ATR | POLH | POLH | FANCL | AT5G65740 |
| MSH6 | MSH6 MSH7 | RAD9A RAD9B | RAD9 | PCNA | PCNA PCNA2 | LIG1 | LIG1 AT1G49250 |
| NEIL2 | FPG1 | CHAF1A | FAS1 | CDC45 | CDC45 | MSH2 | MSH2 |
| NTHL1 | NTH1 NTH2 | POLR21 | NRPB9A NRPB9B | GTF2H2 GTF2H2C | ATGTF2H2 | RPA2 | RPA2A RPA2B |
| MUTYH | MYH | GTF2H1 | TFB1-1 | INO80 | INO80 | CHAF1B | FAS2 |
| PRPF19 | PRP19A | LIG4 | LIG4 | MSH3 | MSH3 | SMC5 | SMC5 |
| FEN1 | FEN1 | RFC1 | RFC1 | GTF2H3 | AT1G18340 | SMARCAD1 | ETL1 |
| RAD17 | RAD17 | PARP1 | PARP1 |
| BER | NER | MMR | HR | NHEJ | |||||
|---|---|---|---|---|---|---|---|---|---|
| Hs | At | Hs | At | Hs | At | Hs | At | Hs | At |
| APEX2 | ARP APE1L APE2 | ERCC3 | XPB2 XPB1 | MSH6 | MSH7 MSH6 | WRN BLM | RECQL4A | PARP2 PARP1 | PARP2 |
| NTHL1 | NTH2 NTH1 | RAD23B RAD23A | RAD23A RAD23B RAD23C RAD23D | RNASEH2A | AT2G25100 | RAD51 DMC1 | RAD51 | XRCC6 | KU70 |
| UNG | ATUNG | ERCC2 | UVH6 | MLH1 | MLH1 | FIGNL1 | AT3G27120 | XRCC5 | KU80 |
| FEN1 | FEN1 | GTF2H2 GTF2H2C | ATGTF2H2 | PCNA | PCNA PCNA2 | RAD54L RAD54B | CHR25 | XRCC1 | XRCC1 |
| MRE11A | MRE11 | POLR2I | NRPB9A NRPB9B | MSH2 | MSH2 | MTOR | TOR | ||
| OGG1 | OGG1 | XPC | ATRAD4 | PMS2 | PMS1 | ERCC4 | UVH1 | ||
| MUTYH | MYH | ERCC5 | UVH3 | MSH5 | MSH5 | ATR | ATR | ||
| MPG | MAG | GTF2H4 | AT4G17020 | MSH4 | MSH4 | GINS2 | GINS2 | ||
| NEIL2 | FPG1 | GTF2H3 | AT1G18340 | MSH3 | MSH3 | SMC5 | SMC5 | ||
| DDB1 | DDB1A DDB1B | MLH3 | MLH3 | ERCC1 | ERCC1 | ||||
| GTF2H1 | TFB1-1 TFB1-3 | GINS4 | SLD5 | ||||||
| LIG4 | LIG4 | MCM8 | MCM8 | ||||||
| POLL | AT1G10520 | MUS81 | MUS81 | ||||||
| CDC45 | CDC45 | ||||||||
| NSMCE1 | emb1379 | ||||||||
| SHFM1 | ATDSS1(V) | ||||||||
| POLD3 | POLD3 | ||||||||
| XRCC3 | XRCC3 | ||||||||
| NSMCE2 | MMS21 | ||||||||
| BRCA2 | BRCA2A BRCA2B | ||||||||
| NBN | NBS1 | ||||||||
| XRCC2 | XRCC2 | ||||||||
| ZSWIM7 | AT4G33925 | ||||||||
| RAD51B | RAD51B | ||||||||
| 1. Mechanism (GO Term) | 2. # Hs Genes | 3. # Hs Genes That Have Orthologues in At | 4. % | 5. # At Genes | 6. # At Genes That Have Orthologues in Hs | 7. % | 8. # Suggested Genes Arisen from Our Analysis for Hs | 9. # Suggested Genes Arisen from Our Analysis for At |
|---|---|---|---|---|---|---|---|---|
| DNA repair GO:0006281 | 507 | 259 | 51.1 | 300 | 185 | 61.7 | 86 | 243 |
| BER GO:0006284 | 52 | 32 | 61.5 | 29 | 12 | 41.4 | 3 | 36 |
| NER GO:0006289 | 124 | 59 | 47.6 | 30 | 21 | 70.0 | 5 | 57 |
| MMR GO:0006298 | 43 | 14 | 32.6 | 17 | 12 | 70.6 | 0 | 1 |
| HR GO:0000724 | 162 | 76 | 46.9 | 50 | 37 | 74.0 | 38 | 87 |
| NHEJ GO:0006303 | 73 | 9 | 12.3 | 7 | 4 | 57.1 | 0 | 8 |
| Total number of ‘DNA repair’ genes associated with “new roles” arisen from this analysis (see Figure 8 and Figure 9) | 95 | 281 | ||||||
| Total number of “entirely new” genes that have arisen from this analysis after the comparison with the “previously established” DNA repair genes | 84 | 234 | ||||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikitaki, Z.; Pavlopoulou, A.; Holá, M.; Donà, M.; Michalopoulos, I.; Balestrazzi, A.; Angelis, K.J.; Georgakilas, A.G. Bridging Plant and Human Radiation Response and DNA Repair through an In Silico Approach. Cancers 2017, 9, 65. https://doi.org/10.3390/cancers9060065
Nikitaki Z, Pavlopoulou A, Holá M, Donà M, Michalopoulos I, Balestrazzi A, Angelis KJ, Georgakilas AG. Bridging Plant and Human Radiation Response and DNA Repair through an In Silico Approach. Cancers. 2017; 9(6):65. https://doi.org/10.3390/cancers9060065
Chicago/Turabian StyleNikitaki, Zacharenia, Athanasia Pavlopoulou, Marcela Holá, Mattia Donà, Ioannis Michalopoulos, Alma Balestrazzi, Karel J. Angelis, and Alexandros G. Georgakilas. 2017. "Bridging Plant and Human Radiation Response and DNA Repair through an In Silico Approach" Cancers 9, no. 6: 65. https://doi.org/10.3390/cancers9060065
APA StyleNikitaki, Z., Pavlopoulou, A., Holá, M., Donà, M., Michalopoulos, I., Balestrazzi, A., Angelis, K. J., & Georgakilas, A. G. (2017). Bridging Plant and Human Radiation Response and DNA Repair through an In Silico Approach. Cancers, 9(6), 65. https://doi.org/10.3390/cancers9060065