1. Introduction
The direct linear plot (DLP) is a graphic method to estimate kinetic constants from enzymatic reactions based on the median as a statistic. The method was developed by Eisenthal and Cornish-Bowden in 1974 [
1]. The accuracy and robustness of this method were tested during the estimation of
Vmax and
Km from the Michaelis–Menten equation [
1,
2]. As in this article, all of the recent studies regarding the application of DLP are based on the estimation of these two parameters from the Michaelis–Menten equation. The application of DLP to equations with more than two parameters has not been studied until now [
3]. This median-based method was applied herein to estimate three kinetic constants from the substrate-uncompetitive inhibition equation. The results indicated that DLP was not only applicable to equations with more than two parameters, but it was reliable and robust when compared to the least-squares (LS) method. In the present article, we explored the application of DLP to the product-competitive inhibition equation, which is a different type of problem, as will be exposed. Some attempts to estimate inhibition constants from enzyme kinetics using DLP have involved estimating apparent kinetic constants and using secondary plots to estimate the values of these inhibition constants. The application of DLP to estimate inhibition constants was first proposed by Eisenthal and Cornish-Bowden [
1]. The literature contains outdated examples of this application because the use of DLP to estimate inhibition constants in one-stage has not been assessed [
4,
5,
6,
7,
8]. In the literature, DLP has been used to estimate the apparent kinetic constants
Vmax and
Km; however, studies have used secondary plots to estimate the inhibition constant
Kp. The present article studied the application of DLP to estimate the product inhibition constant in one-stage treatment, avoiding the estimation of apparent kinetic constants and secondary plots. As the proper characterization of inhibition in pharmaceutical and biotechnological applications is a major concern, the importance of this proposal is that it opens up the possibility of using a reliable and robust method to estimate product inhibition constants. The purpose of this study is to estimate
Vmax,
Km, and
Kp from the competitive inhibition equation in just one stage of calculations, avoiding the apparent constants and secondary plots. This means the values of the product inhibition constants must be obtained from the DLP method, which has never been tried before. The problem is outlined as follows.
The equation for competitive product inhibition involves three parameters—
Vmax,
Km, and
Kp—and three variables—substrate concentration, product concentration, and initial reaction rate (Equation (1)).
This is a different type of problem compared to previous articles where the substrate-uncompetitive inhibition equation involved three parameters—
Vmax,
Km, and
KS—but only two variables—substrate concentration and initial reaction rate (Equation (2)) [
3].
The experimental dataset required is traditionally used in the initial reaction rate method, which denotes a series of initial rates vs. substrate concentrations at constant product concentrations, as indicated in the scheme of
Figure 1.
Equation (1) can be rearranged to Equation (3) in the following form:
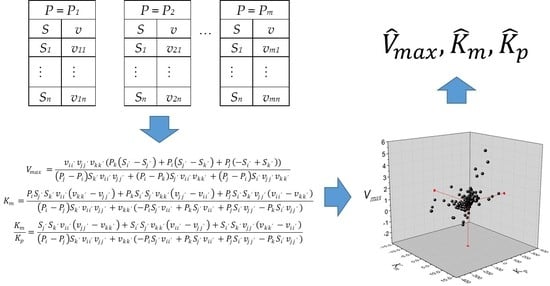
The definition of
vij is the rate of product concentration
Pi and substrate concentration
Sj. As the estimation of three parameters is required, three data pairs and three equations are needed to solve the matrix system in Equation (4).
Likewise, the definition of
vii’ =
v(
Pi,
Si’), i.e.,
i values are between 1 and
m (the number of product concentrations), and
i’ values are between 1 and
n (the number of substrate concentrations). Alternatively, the solution for Equation (4) is presented in Equation (5) in algebraic form.
The rank of the matrix in Equation (4) must be 3 for the system to have a solution. For the case when three product concentrations are taken from the same set, Pi = Pj = Pk, the system has no solution because the rank of the resulting matrix is 2. This problem can be easily checked by observing the denominators of Equation (5) when replacing Pi, Pj, and Pk with the same value of product concentration. The strategy proposed to solve this system is to take at least two different values of product concentration. In this way, the total number of feasible data combinations will be calculated using Equation (6).
In consequence, calculations of the kinetic constants
Vmax,
Km, and
Kp will be obtained from the combination of two
P values from the same dataset, one
P from a different one, and the combination of three different
P values from three datasets. A list of
Vmax,
Km, and
Kp values from the total combinations indicated in Equation (6) will be obtained. Finally, the estimators of
Vmax,
Km, and
Kp correspond to the median values from the list.
This article aims to compare the quality of the kinetic constant estimations from the product inhibition equation between the DLP and the LS methods.
2. Results and Discussion
As in our previous article [
3], this problem consists of estimating three kinetic constants, but in this new case, three experimental variables will be used. Experimental data of initial rate perturbed with a random error of variance
σ2 were simulated in a dataset of 1000 runs. The dataset, according to
Table 1, considers the values of the kinetic constants 1, 1 and 10 for
Vmax,
Km, and
Kp, respectively, and the initial rates vs. the substrate concentrations at different product concentrations were plotted in
Figure 2.
These data were used to estimate the kinetic constants using the DLP and LS methods. An example of the dataset obtained for the values of the kinetic constants
Vmax,
Km, and
Kp with a normal distribution of error and variance 0.01 is plotted in
Figure 3.
The number of kinetic constant values plotted in
Figure 3 and
Figure 4 was 6370. This amount of data was obtained avoiding the use of the same three product concentrations during the calculations with Equation (4). The dispersion of the data was enormous compared to the value of the kinetic constant, as can be observed in
Figure 4. This is a typical result of the application of the DLP method to the estimation of kinetic constants. This behavior can be observed in previous publications [
1,
3,
9]. Many points were left out of the layers just for illustration purposes; however, the vast majority of data are inside of layers corresponding to 98%, 99% and 99% for
Vmax,
Km and
Kp, respectively. In terms of dispersion, 56% of calculated
Vmax values are in the range 0.95–1.05; 22% of calculated Km values are in the range 0.95–1.05 and 24% of calculated
Kp values are in the range 9.5–10.5. The amount of data plotted in
Figure 3 and
Figure 4 was obtained to estimate the kinetic constants
Vmax,
Km, and
Kp by calculating the median for each of them. This is the procedure needed in routine experiments to determine the inhibition constant
Kp. The statistical evaluation of DLP requires the repetition of this procedure to avoid bias and erroneous conclusions. A total of 1000 experimental runs were carried out to evaluate and compare the quality of the estimations between the DLP and the LS methods. A comparison of the lower values of the sum of squared residuals (
SSR) between DLP and LS is plotted in
Figure 5. The DLP method obtained lower values of
SSR for all the values of variance in the range from 0.001 to 0.020 when both procedures—calculation of the median of
Km/
Kp and the median of
Kp—were carried out. The calculation of the median of
Km/
Kp was slightly superior for almost all variances explored. For example, at variance 0.010, the calculation of the median of
Km/
Kp obtained a frequency of 0.8 compared to 0.78 for calculation of the median of
Kp. This was not significant to conclude that one procedure was better than the other. However, when both calculations were compared regarding the
SSR between them, the result depended on the variance, as shown in
Figure 6a. The calculation of the median of
Km/
Kp obtained a lower
SSR at variance values lower than 0.010. The calculation of the median of
Kp offered lower values of
SSR at variances higher than 0.010. The effect of the ratio
Kp/
Km was slightly superior to the calculation of the median of
Kp, as shown in
Figure 6b. However, this was not significantly different. In consequence, which procedure is better, in terms of the
SSR, depends on the experimental error.
The relative error of the estimated kinetic constants by LS and DLP methods was plotted for 1000 experimental runs in
Figure 7. The DLP always resulted in a lower accumulation of relative error during estimations of the three kinetic constants. The product inhibition constant
Kp accumulated the highest relative error when estimated by both LS and DLP methods. However, the accumulated relative error was less than half with DLP compared to LS. This result shows that DLP is a very accurate method to estimate the competitive inhibition constant. The product inhibition constant
Kp estimated with DLP method accumulated a lower relative error than the LS method even at higher variance
σ2 (experimental error), as shown in
Figure 8. The behavior patterns of both the median of
Km/
Kp and the median of
Kp were repeated. The median of
Km/
Kp was slightly more accurate at variances lower than 0.010. At higher values, the median of
Kp was more accurate.
The effect of outliers was analyzed by adding fixed error values to the initial rate
v34 (
S = 1;
P = 10). The added error ranged from −0.01 to +0.01. The results are shown in
Figure 9. The estimated values of
Vmax by LS were constant due to an artifact of the estimation method. The estimated
Vmax was obtained by choosing the highest value from the five non-linear regressions corresponding to the five datasets of product concentrations. The error was added to the initial rate value corresponding to the third dataset of product concentration; in consequence, the highest value of
Vmax was not perturbed, and it was always the same value. The DLP method estimated
Km and the
Kp with more accuracy and with less perturbation than the LS method. Although the LS method estimated
Kp with more accuracy at some high positive values of fixed error, the results clearly showed that DLP was more robust than the LS method. The perturbation generated by the added error is plotted in
Figure 10. The perturbation of Vmax was omitted due to the artifact during its estimation by the LS method. The results indicated that the estimation of kinetic constants
Km and
Kp was much less perturbed when the DLP method was used. Almost all the perturbations were maintained below 1%, compared to the LS method, where perturbations were in the range from 4% to 8%.
The DLP method exhibited the same pattern of behavior than that observed during application to the uncompetitive inhibition equation [
3]. Lower
SSR and estimation errors compared to the LS method were observed during estimations of the competitive inhibition constant. Again, the DLP method demonstrated accuracy and robustness during the estimation of kinetic constants. The possibility of using the DLP method to estimate inhibition constants in pharmaceutical and biotechnological studies was demonstrated. The performance of the DLP method applied to the competitive inhibition equation was different compared to the application of the substrate-uncompetitive inhibition equation previously studied. The addition of a variable product concentration caused mathematical indetermination during the calculations. The problem was overcome by avoiding using the same three product concentrations in Equations (4) and (5). This obligated a reduction in the number of combinations from 6545 to 6370, where 175 combinations resulted in the indetermination of the matrix (Equation (4)). Another difference is the dependence of
Kp on the value of
Km, which can be observed in Equation (5), where the calculation consisted of the
Km/
Kp value, and
Kp was calculated after obtaining
Km. A significant implication of this is the transmission of the
Km error to
Kp. Interestingly, based on the observations, this effect was not important. In general, the estimation of
Kp by DLP was more accurate and robust than estimation by the LS method. The application of DLP to the uncompetitive inhibition equation exhibited a higher accuracy just in some cases compared to the LS method. In terms of robustness, the DLP was highly superior to the LS method. Presently, in this study, a higher superiority in terms of accuracy and robustness was observed. Despite the higher complexity of the competitive inhibition equation, requiring one more independent variable than the substrate-uncompetitive inhibition equation, the results were better. The transmission of error from
Km to
Kp could be the key. This error is surely amplified when transmitted from one constant to another, and based on the observations, the effect was better softened by DLP compared to the LS method.
As Cornish-Bowden and Endrenyi commented [
10], the median method cannot readily be generalized to equations of more than two parameters. This sentence became true when we found a mathematical issue during the application of the DLP to the product’s inhibition equation, where an indeterminate system of equations (Equation (5)) is generated when the same value for the product concentration was used. However, this problem can be solved by selecting the proper combination of data. We, again, demonstrated that DLP can be applied to equations with more than two parameters. Presently, we also demonstrated the application to equations with more than two variables.
We conclude that the DLP method is better than the LS method to estimate the product’s competitive inhibition constant in terms of accuracy and robustness. We now count on a new tool to estimate the product-competitive inhibition constant with reliable results.