The Current State of Research on PET Hydrolyzing Enzymes Available for Biorecycling
Abstract
:1. Introduction
2. Comparison of Thermophilic and Mesophilic Cutinases: Desirable Characteristics for the Enzymatic Recycling of Polyethylene Terephthalate (PET)
3. Structural Insights into the Reaction Mechanism of PET-Hydrolyzing Enzymes
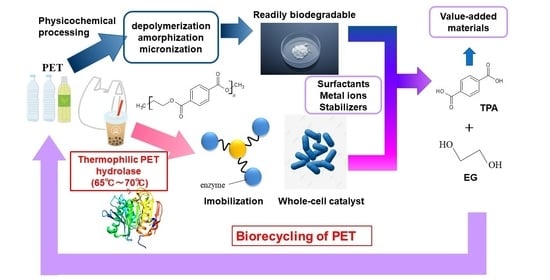
4. Perspectives for Biorecycling of PET
5. Concluding Remarks
Funding
Data Availability Statement
Conflicts of Interest
References
- Geyer, R.; Jambeck, J.R.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PlasticsEurope. Plastics—The Facts 2019. Available online: https://www.plasticseurope.org/application/files/9715/7129/9584/FINAL_web_version_Plastics_the_facts2019_14102019.pdf (accessed on 12 January 2021).
- World Economic Forum. The New Plastics Economy: Rethinking the Future of Plastics. 2016. Available online: http://www3.weforum.org/docs/WEF_The_New_Plastics_Economy.pdf (accessed on 12 January 2021).
- Müller, R.-J.; Schrader, H.; Profe, J.; Dresler, K.; Deckwer, W.-D. Enzymatic degradation of poly(ethylene terephthalate): Rapid hydrolysis using a hydrolase from T. fusca. Macromol. Rapid Commun. 2005, 26, 1400–1405. [Google Scholar] [CrossRef]
- Thumarat, U.; Nakamura, R.; Kawabata, T.; Suzuki, H.; Kawai, F. Biochemical and genetic analysis of a cutinase-type polyesterase from a thermophilic Thermobifida alba AHK119. Appl. Microbiol. Biotechnol. 2012, 95, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Herrero Acero, E.; Ribitsch, D.; Steinkellner, G.; Gruber, K.; Greimel, K.; Eiteljoerg, I.; Trotscha, E.; Wei, R.; Zimmermann, R.; Zinn, M.; et al. Enzymatic surface hydrolysis of PET: Effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules 2011, 44, 4632–4640. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Oeser, T.; Kühn, N.; Barth, M.; Schmidt, J.; Zimmermann, W. Functional characterization and structural modeling of synthetic polyester-degrading hydrolases form Thermomonospora curvata. AMB Express 2014, 4, 44. [Google Scholar] [CrossRef] [Green Version]
- Kawai, F.; Oda, M.; Tamashiro, T.; Waku, T.; Tanaka, N.; Yamamoto, M.; Mizushima, H.; Miyakawa, T.; Tanokura, M. A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 2014, 98, 10053–10064. [Google Scholar] [CrossRef]
- Sulaiman, S.; Yamato, S.; Kanaya, E.; Kim, J.-J.; Koga, Y.; Takano, K.; Kanaya, S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012, 78, 1556–1562. [Google Scholar] [CrossRef] [Green Version]
- Ronqvist, Å.M.; Venchun, X.; Lu, W.; Gross, R.A. Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules 2009, 42, 5128–5138. [Google Scholar] [CrossRef]
- Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K. A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351, 1196–1199. [Google Scholar] [CrossRef]
- eLetters. Available online: https://science.sciencemag.org/content/351/6278/1196/tab-e-letters (accessed on 2 February 2021).
- Wei, R.; Song, C.; Gräsing, D.; Schneider, T.; Bielytskyi, P.; Böttche, D.; Matysik, J.; Bornscheuer, U.T.; Zimmerman, W. Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Nat. Commun. 2019, 10, 5581. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.-L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Kawai, F.; Kawabata, T.; Oda, M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 2019, 103, 4253–4268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, F.; Kawabata, T.; Oda, M. Current state and perspectives related to the polyethylene terephthalate hydrolases available for biorecycling. ACS Sustain. Chem. Eng. 2020, 8, 8894–8908. [Google Scholar] [CrossRef]
- Joo, S.; Cho, I.J.; Seo, H.; Son, H.F.; Sagong, H.Y.; Shin, T.J.; Choi, S.Y.; Lee, S.Y.; Kim, K.J. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat. Commun. 2018, 9, 382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.; Hazen, T.; Streit, W.R. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, H.; Shigeno-Akutsu, Y.; Nomura, N.; Nakahara, T.; Nakajima-Kambe, T. Cloning and sequence analysis of poly(tetramethylene succinate) depolymerase from Acidovorax delafieldii strain BS-3. J. Biosci. Bioeng. 2002, 93, 245–247. [Google Scholar] [CrossRef]
- Almeida, E.L.; Rincó, A.F.C.; Jackson, S.A.; Dobson, A.D.W. In silico screening and heterogous expression of a polyethylene terephthalate hydrolase (PETase)-like enzyme (SM14est) with polycaprolactone(PCL)-degrading activity, from the marine sponge-derived strain Streptomyces sp. SM14. Front. Microbiol. 2019, 10, 2187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bollinger, A.; Thies, S.; Knieps-Grűnhagen, E.; Gertzen, C.; Kobus, S.; Hőppner, A.; Ferrer, M.; Gohlke, H.; Smits, S.H.J.; Jaeger, K.-E. A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri—Structural and functional insights. Front. Microbiol. 2020, 11, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, X.; Ni, K.; Hao, H.; Shang, Y.; Zhao, B.; Qian, Z. Secretory expression in Bacillus subtilis and biochemical characterization of a highly thermostable polyethylene terephthalate hydrolase from bacterium HR29. Enzym. Microb. Technol. 2021, 143, 109715. [Google Scholar] [CrossRef]
- Kato, S.; Sakai, S.; Hirai, M.; Tasumi, E.; Nishizawa, M.; Suzuki, K.; Takai, K. Long-term cultivation and metagenomics reveal ecophysiology of previously uncultivated thermophiles involved in biogeochemical nitrogen cycle. Microbes Environ. 2018, 33, 107–110. [Google Scholar] [CrossRef] [Green Version]
- Son, H.F.; Cho, I.J.; Joo, S.; Seo, H.; Sagong, H.; Choi, S.Y.; Lee, S.Y.; Kim, K. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for highly efficient PET degradation. ACS Catal. 2019, 9, 3519–3526. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, Y.; Liu, X.; Dong, S.; Tian, Y.; Qiao, Y.; Han, J.; Li, C.; Han, X.; Liu, W.; et al. Computational redesign of PETase for plastic biodegradation by GRAPE strategy. bioRxiv 2019. [Google Scholar] [CrossRef]
- Wei, R.; Oeser, T.; Schmidt, J.; Meier, R.; Barth, M.; Then, J.; Zimmermann, W. Engineered bacterial polyester hydrolases efficiently degrade polyethylene terephthalate due to relieved product inhibition. Biotechnol. Bioeng. 2016, 113, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, S.; You, D.-J.; Kanaya, E.; Koga, Y.; Kanaya, S. Crystal structure and thermodynamic and kinetic stability of metagenome-derived LC-cutinase. Biochemistry 2014, 53, 1858–1869. [Google Scholar] [CrossRef] [PubMed]
- Oda, M.; Yamagami, Y.; Inaba, S.; Oida, T.; Yamamoto, M.; Kitajima, S.; Kawai, F. Enzymatic hydrolysis of PET: Functional roles of three Ca2+ ions bound to a cutinase-like enzyme, Cut190*, and its engineering for improved activity. Appl. Microbiol. Biotechnol. 2018, 102, 10067–10077. [Google Scholar] [CrossRef]
- Thumarat, U.; Kawabata, T.; Nakajima, M.; Nakajima, M.; Sugiyama, A.; Yazaki, K.; Tada, T.; Waku, T.; Tanaka, N.; Kawai, F. Comparison of genetic structures and biochemical properties of tandem cutinase-type polyesterases from Thermobifida alba AHK119. J. Biosci. Bioeng. 2015, 120, 491–497. [Google Scholar] [CrossRef]
- Ribitsch, D.; Acero, E.H.; Greimel, K.; Eiteljoerg, I.; Trotscha, E.; Freddi, G.; Schwab, H.; Guebitz, G.M. Characterization of a new cutinase from Thermobifida alba for PET-surface hydrolysis. Biocatal. Biotransform. 2012, 30, 2–9. [Google Scholar] [CrossRef]
- Acero, E.H.; Ribitsch, D.; Dellacher, A.; Zitzenbacher, S.; Marold, A.; Steinkellner, G.; Gruber, K.; Schwab, H.; Guebitz, G.M. Surface engineering of a cutinase from Thermobifida cellulosilytica for improved polyester hydrolysis. Biotechnol. Bioeng. 2013, 110, 2581–2590. [Google Scholar] [CrossRef]
- Ribitsch, D.; Acero, E.H.; Greimel, K.; Dellacher, A.; Zitzenbacher, S.; Marold, A.; Rodriguez, R.D.; Steinkellner, G.; Gruber, K.; Schwab, H.; et al. A new esterase from Thermobifida halotolerans hydrolyses polyethylene terephthalate (PET) and polylactic acid (PLA). Polymers 2012, 4, 617–629. [Google Scholar] [CrossRef]
- Wei, Y.; Swenson, L.; Casdtro, C.; Derewenda, U.; Minor, W.; Arai, H.; Aoki, J.; Inoue, K.; Servin-Gonzalez, L.; Derewenda, Z.S. Structure of a microbial homologue of mammalian platelet-activating factor acetylhydrolases: Streptomyces exfoliates lipase at 1.9 Å resolution. Structure 1998, 6, 511–519. [Google Scholar] [CrossRef] [Green Version]
- Kitadokoro, K.; Thumarat, U.; Nakamura, R.; Nishimura, K.; Karatani, H.; Suzuki, H.; Kawai, F. Crystal structure of cutinase Est119 from Thermobifida alba AHK119 that can degrade modified polyethylene terephthalate at 1.76 Å resolution. Polym. Degrad. Stab. 2012, 97, 771–775. [Google Scholar] [CrossRef]
- Roth, C.; Wei, R.; Oeser, T.; Then, J.; Foellner, C.; Zimmermann, W.; Straeter, N. Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca. Appl. Microbiol. Biotechnol. 2014, 98, 7815–7823. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, T.; Mizushima, H.; Ohtsuka, J.; Oda, M.; Kawai, F.; Tanokura, M. Structural basis for the Ca2+-enhanced thermostability and activity of PET-degrading cutinase from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 2015, 99, 4297–4307. [Google Scholar] [CrossRef] [PubMed]
- Numoto, N.; Kamiya, N.; Bekker, G.J.; Yamagami, Y.; Inaba, S.; Ishii, K.; Uchiyama, S.; Kawai, F.; Ito, N.; Oda, M. Structural dynamics of the PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190 in substrate-bound states elucidates the Ca2+-driven catalytic cycle. Biochemistry 2018, 57, 5289–5300. [Google Scholar] [CrossRef] [PubMed]
- Ribitsch, D.; Hromic, A.; Zitzenbacher, S.; Zartl, B.; Gamerith, C.; Pellis, A.; Jungbauer, A.; Lyskowski, A.; Steinkellner, G.; Gruber, K.; et al. Small cause, large effect: Structural characterization of cutinases from Thermobifida cellusilytica. Biotechnol. Bioeng. 2017, 114, 2481–2488. [Google Scholar] [CrossRef] [PubMed]
- Then, J.; Wei, R.; Oeser, T.; Barth, M.; Belisario-Ferrari, M.R.; Schmidt, J.; Zimmermann, W. Ca2+ and Mg2+ binding site engineering increases the degradation of polyethylene terephthalate films by polyester hydrolase from Thermobifida fusca. Biotechnol. J. 2015, 10, 592–598. [Google Scholar] [CrossRef]
- Then, J.; Wei, R.; Oeser, T.; Gerdts, A.; Schmidt, J.; Barth, M.; Zimmermann, W. A disulfide bridge in the calcium binding site of a polyester hydrolase increases its thermal stability and activity against polyethylene terephthalate. FEBS Open Bio 2016, 6, 425–432. [Google Scholar] [CrossRef] [Green Version]
- Kitadokoro, K.; Kakara, M.; Matsui, S.; Osokoshi, R.; Thumarat, U.; Kawai, F.; Kamitani, S. Structural insights into the unique polylactate-degrading mechanism of Thermobifida alba cutinase. FEBS J. 2019, 286, 2087–2098. [Google Scholar] [CrossRef]
- Austin, H.P.; Allen, M.D.; Donohoe, B.S.; Rorrer, N.A.; Kearns, F.L.; Silveria, R.L.; Polland, B.C.; Dominick, G.; Duman, R.; Omari, K.E.; et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. USA 2018, 115, E4350–E4357. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.C.; Han, X.; Ko, T.-P.; Liu, W.; Guo, R.-T. Structural studies reveal the molecular mechanism of PETase. FEBS J. 2018, 285, 3717–3723. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; He, L.; Wang, L.; Li, T.; Li, C.; Liu, H.; Luo, Y.; Bao, R. Protein crystallography and site-direct mutagenesis analysis of the poly(ethylene terephthalate) hydrolase PETase from Ideonella sakaiensis. ChemBioChem 2018, 19, 1471–1475. [Google Scholar] [CrossRef] [PubMed]
- Zumstein, M.T.; Rechsteiner, D.; Roduner, N.; Perz, V.; Ribitsch, D.; Guebitz, G.M.; Kohler, H.-P.E.; McNeill, K.; Sander, M. Enzymatic hydrolysis of polyester thin films at the nanoscale: Effects of polyester structure and enzyme active-site accessibility. Environ. Sci. Technol. 2017, 51, 7476–7485. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Breite, D.; Song, C.; Gräsing, D.; Ploss, T.; Hille, P.; Schwerdtfeger, R.; Matysik, J.; Schultze, A.; Zimmermann, W. Biocatalytic degradation efficiency of postconsumer polyethylene terephthalate packaging determined by their polymer microstructures. Adv. Sci. 2019, 6, 1900491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirke, A.; White, C.; Englaender, J.A.; Zwarycz, A.; Butterfoss, G.L.; Linhardt, R.J.; Gross, R.A. Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: Mechanism and effect on PET hydrolysis. Biochemistry 2018, 57, 1190–1200. [Google Scholar] [CrossRef]
- Yan, F.; Wei, R.; Cui, Q.; Bornscheuer, U.T.; Liu, Y.-J. Thermophilic whole-cell degradation of polyethylene terephthalate using engineered Clostridium thermocellum. Microb. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Barth, M.; Honak, A.; Oeser, T.; Wei, R.; Belisário-Ferrari, M.R.; Then, J.; Schmidt, J.; Zimmermann, W. A dual enzyme system composed of a polyester hydrolase and a carboxylesterase enhances the biocatalytic degradation of polyethylene terephthalate films. Biotechnol. J. 2016, 11, 1082–1087. [Google Scholar] [CrossRef]
- Carniel, A.; Voloni, É.; Junior, J.N.; da Conceição Gomes, A.; de Castro, A.M. Lipase from Candida antarctica (CALB) and cutinase from Humicola insolens act synergetically for PET hydrolysis to terephthalic acid. Process. Biochem. 2017, 59, 84–90. [Google Scholar] [CrossRef]

| Enzyme | Source | PET Degradation | Product Level | PET Used | Reaction Conditions | Reference |
|---|---|---|---|---|---|---|
| Thermophilic cutinase | ||||||
| TfH (BTA-1) | Thermobifida fusca DSM43793 | ≈50% | amorphous PET film or amorphous bottle | 55 °C, 3 weeks | [4] | |
| TfCut2 variant | T. fusca KW3 | 42% | 49 mM | PET-GF film a | 65 °C, 50 h | [26] |
| LC-cutinase | Metagenome from leaf branch compost | ≤25% | 28 mM | PET package b | 70 °C, 24 h | [27] |
| LC-cutinase variant | Metagenome from leaf branch compost | 90% | 936 mM | amorphized and micronized PET | 72 °C, 10 h | [14] |
| BhrPETase | thermophilic bacterium strain HR29 | 6.3 mM | PET powder | 70 °C, 20 h | [22] | |
| Cut190 variant | Saccharomonospora viridis AHK190 | 34% 59% | 15 mM 29 mM | PET-GF film a PET package b | 70 °C, 3 days 60 °C, 3 days | [28] |
| HiC (Novo) | Humicola insolens | 97% | 135 mM | PET-GF film a | 70 °C, 4 days | [10] |
| Est 1 | T. alba AHK119 | Not confirmed | [29] | |||
| Tha_Cut1 | T. alba DSM43185 | Not confirmed | [30] | |||
| Thc_Cut2 variant | T. cellusilytica DSM44535 | 0.45 mM | PET film (not specified) | 50 °C, 2 days | [31] | |
| Tcur1278 | T. curvata DSM43183 | PET nanoparticles | 50–60 °C | [7] | ||
| Thh_Est | T. halotolerans DSM44931 | Not confirmed | [32] | |||
| Mesophilic cutinases | ||||||
| IsPETase | Ideonella sakaiensis | 0.23% | 0.3 mM | amorphous PET film | 30 °C, 48 h | [11] |
| IsPETase DuraPETase (IsPETase variant) | Ideonella sakaiensis | 12 µM 1.8 mM 3.4 mM | PET film | 37 °C, 12 h d 40 °C, 72 h 40 °C, 10 days | [25] | |
| PBS depolymerase | Acidovorax delafieldii strain BS-3 | Not confirmed | 30 °C | [19] | ||
| SM14est c | Streptomyces sp. SM14 (marine) | Not confirmed | 28 °C | [20] | ||
| PE-H PE-H/Y250S | Pseudomonas aestusnigri (marine) | 21 µM 31 µM | PET-GF film a | 30 °C, 48 h | [21] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawai, F. The Current State of Research on PET Hydrolyzing Enzymes Available for Biorecycling. Catalysts 2021, 11, 206. https://doi.org/10.3390/catal11020206
Kawai F. The Current State of Research on PET Hydrolyzing Enzymes Available for Biorecycling. Catalysts. 2021; 11(2):206. https://doi.org/10.3390/catal11020206
Chicago/Turabian StyleKawai, Fusako. 2021. "The Current State of Research on PET Hydrolyzing Enzymes Available for Biorecycling" Catalysts 11, no. 2: 206. https://doi.org/10.3390/catal11020206
APA StyleKawai, F. (2021). The Current State of Research on PET Hydrolyzing Enzymes Available for Biorecycling. Catalysts, 11(2), 206. https://doi.org/10.3390/catal11020206