Improving Effects of Laccase-Mediated Pectin–Ferulic Acid Conjugate and Transglutaminase on Active Peptide Production in Bovine Lactoferrin Digests
Abstract
:1. Introduction
2. Results
2.1. Compositions and Characteristic Ratios of PP and PF
2.2. NMR-Evidenced Structural Features of PP and PF
2.3. Particle Size Distributions of bLf, LfPF Complex, and LfPFTG Complex
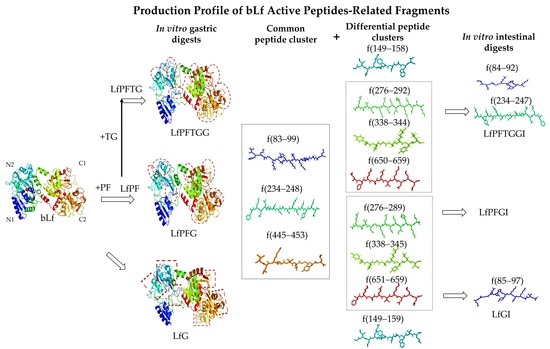
2.4. Differential Peptide Distributions after In Vitro Gastrointestinal Digestion
2.5. Differences in Active Peptide Fragments between Samples
2.6. bLf Active Peptide Changes during In Vitro Gastrointestinal Digestion
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Preparation of PP-FA Conjugate (PF)
4.3. Preparation of bLf-PF Complex (LfPF)
4.4. Measurement of Uronic Acid Content
4.5. Determination of Total Phenolic Content
4.6. Measurement on Degree of Esterification
4.7. Assessment of Monosaccharide Composition
4.8. Nuclear Magnetic Resonance (NMR) Analysis
4.9. Examination of Particle Size Distribution
4.10. ζ-Potential Analysis
4.11. Digestomics Analysis
4.11.1. In Vitro Simulated Gastrointestinal Digestion
4.11.2. Peptide Purification
4.11.3. UPLC-MS-MS Analysis for Peptide Distribution
4.11.4. Homology Matching and Molecular Modeling
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iglesias-Figueroa, B.F.; Espinoza-Sánchez, E.A.; Siqueiros-Cendón, T.S.; Rascón-Cruz, Q. Lactoferrin as a nutraceutical protein from milk, an overview. Int. Dairy J. 2019, 89, 37–41. [Google Scholar] [CrossRef]
- Elzoghby, A.O.; Abdelmoneem, M.A.; Hassanin, I.A.; Abd Elwakil, M.M.; Elnaggar, M.A.; Mokhtar, S.; Fang, J.-Y.; Elkhodalry, K.A. Lactoferrin, a multi-functional glycoprotein: Active therapeutic, drug nanocarrier & targeting ligand. Biomaterials 2020, 263, 120355. [Google Scholar] [PubMed]
- Ji, Y.; Ai, L.Z.; Xing, M.X.; Xie, F.; Lai, P. Advances in research on bovine lactoferrin-based technical innovation and application. Food Ferment. Ind. 2022, 48, 332–340. [Google Scholar] [CrossRef]
- Agwa, M.M.; Sabra, S. Lactoferrin coated or conjugated nanomaterials as an active targeting approach in nanomedicine. Int. J. Biol. Macromol. 2021, 167, 1527–1543. [Google Scholar] [CrossRef]
- Furlund, C.B.; Ulleberg, E.K.; Devold, T.G.; Flengsrud, R.; Jacobsen, M.; Sekse, C.; Holm, H.; Vegarud, G.E. Identification of lactoferrin peptides generated by digestion with human gastrointestinal enzymes. J. Dairy Sci. 2013, 96, 75–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, M.; Xu, S.; Xu, Z.; Cheng, S.; Wu, D.; Liu, H.; Du, M. Identification of dual-function bovine lactoferrin peptides released using simulated gastrointestinal digestion. Food Biosci. 2021, 39, 100806. [Google Scholar] [CrossRef]
- Shi, P.; Liu, M.; Fan, F.; Chen, H.; Yu, C.; Lu, W.; Du, M. Identification and mechanism of peptides with activity promoting osteoblast proliferation from bovine lactoferrin. Food Biosci. 2018, 22, 19–25. [Google Scholar] [CrossRef]
- Shi, P.; Fan, F.; Chen, H.; Xu, Z.; Cheng, S.; Lu, W.; Du, M. A bovine lactoferrin–derived peptide induced osteogenesis via regulation of osteoblast proliferation and differentiation. J. Dairy Sci. 2020, 103, 3950–3960. [Google Scholar] [CrossRef]
- Ruiz-Giménez, P.; Salom, J.B.; Marcos, J.F.; Vallés, S.; Martínez-Maqueda, D.; Recio, I.; Torregrosa, G.; Alborch, E.; Manzanares, P. Antihypertensive effect of a bovine lactoferrin pepsin hydrolysate: Identification of novel active peptides. Food Chem. 2012, 131, 266–273. [Google Scholar] [CrossRef]
- Picariello, G.; Mamone, G.; Nitride, C.; Addeo, F.; Peranti, P. Protein digestomics: Integrated platforms to study food-protein digestion and derived functional and active peptides. TrAC-Trend Anal. Chem. 2013, 52, 120–134. [Google Scholar] [CrossRef]
- Basak, S.; Annapure, U.S. Trends in “green” and novel methods of pectin modification—A review. Carbohydr. Polym. 2022, 278, 118967. [Google Scholar] [CrossRef]
- Li, D.-q.; Li, J.; Dong, H.-l.; Li, X.; Zhang, J.-q.; Ramaswamy, S.; Xu, F. Pectin in biomedical and drug delivery applications: A review. Int. J. Biol. Macromol. 2021, 185, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Bengoechea, C.; Jones, O.G.; Guerrero, A.; McClements, D.J. Formation and characterization of lactoferrin/pectin electrostatic complexes: Impact of composition, pH and thermal treatment. Food Hydrocoll. 2011, 25, 1227–1232. [Google Scholar] [CrossRef]
- Niu, Z.; Loveday, S.M.; Barbe, V.; Thielen, I.; He, Y.; Singh, H. Protection of native lactoferrin under gastric conditions through complexation with pectin and chitosan. Food Hydrocoll. 2019, 93, 120–130. [Google Scholar] [CrossRef]
- Ji, Y. Study on Preparation Technology, Formulation Optimization and Active Peptide of Lactoferrin-Pectin Complex. Master Thesis, University of Shanghai for Science & Technology, Shanghai, China, 2022. [Google Scholar]
- Karaki, N.; Aljawish, A.; Muniglia, L.; Humeau, C.; Jasniewski, J. Physicochemical characterization of pectin grafted with exogenous phenols. Food Hydrocoll. 2016, 60, 486–493. [Google Scholar] [CrossRef]
- Karaki, N.; Aljawish, A.; Muniglia, L.; Bouguet-Bonnet, S.; Leclerc, S.; Paris, C.; Jasniewski, J. Functionalization of pectin with laccase-mediated oxidation products of ferulic acid. Enz. Microbial Technol. 2017, 104, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Robert, B.; Chenthamara, D.; Subramaniam, S. Fabrication and biomedical applications of arabinoxylan, pectin, chitosan, soyprotein, and silk fibroin hydrogels via laccase—Ferulic acid redox chemistry. Int. J. Biol. Macromol. 2022, 201, 539–556. [Google Scholar] [CrossRef]
- Gao, F.; Ai, L.Z.; Wu, Y.; Lai, F.X.; Zhang, H.; Xie, F.; Song, Z.B. Structural mechanism and structure-antioxidant activity relationship of low-methoxyl pectin-caffeic acid conjugate. Food Sci. 2022, 43, 19–29. [Google Scholar]
- Gao, F.; Ding, N.; Ai, L.; Lai, P.; Zhang, H.; Song, Z. Molecular characteristics, in vitro antioxidant and immunological activities of high-methoxy pectin-phenolic acid derivatives from passion fruit. Food Sci. 2022, 43, 84–94. Available online: https://www.spkx.net.cn/article/2022/1002-6630/2022-43-17-010.html (accessed on 20 September 2022).
- Miwa, N. Innovation in the food industry using microbial transglutaminase: Keys to success and future prospects. Anal. Biochem. 2020, 597, 113638. [Google Scholar] [CrossRef]
- Xia, T.; Gao, Y.; Liu, Y.; Wei, Z.; Xue, C. Lactoferrin particle assembled via transglutaminase-induced crosslinking: Utilization in oleogel-based Pickering emulsions with improved curcumin bioaccessibility. Food Chem. 2022, 374, 131779. [Google Scholar] [CrossRef] [PubMed]
- Martini, S.; Solieri, L.; Tagliazucchi, D. Peptidomics: New trends in food science. Curr. Opin. Food Sci. 2021, 39, 51–59. [Google Scholar] [CrossRef]
- Lin, Y.; An, F.; He, H.; Geng, F.; Song, H.; Huang, Q. Structural and rheological characterization of pectin from passion fruit (Passiflora edulis f. flavicarpa) peel extracted by high-speed shearing. Food Hydrocoll. 2021, 114, 106555. [Google Scholar] [CrossRef]
- Cui, L.; Wang, J.; Huang, R.; Tan, Y.; Zhang, F.; Zhou, Y.; Sun, L. Analysis of pectin from Panax ginseng flower buds and their binding activities to galactin-3. Int. J. Biol. Macromol. 2019, 128, 459–467. [Google Scholar] [CrossRef]
- Liu, D.; Zhai, L.-Y.; Shi, Z.-H.; Hong, H.-L.; Liu, L.-Y.; Zhao, S.-R.; Hu, Y.-B. Purification and fine structural analysis of pectin polysaccharides from Osmunda Japonica Thunb. J. Mol. Struct. 2022, 1269, 133828. [Google Scholar] [CrossRef]
- De Cicco, M.; Mamone, G.; Di Stasie, L.; Ferranti, P.; Addeo, F.; Picariello, G. Hidden “digestome”: Current analytical approaches provide incomplete peptide inventories of food digests. J. Agr. Food Chem. 2019, 67, 7775–7782. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef]
- García-Tejedor, A.; Castelló-Ruiz, M.; Gimeno-Alcañíz, J.V.; Manzanares, P.; Salom, J.B. In vivo antihypertensive mechanism of lactoferrin-derived peptides: Reversion of angiotensin I- and angiotensin II-induced hypertension in Wistar rats. J. Funct. Foods 2015, 15, 294–300. [Google Scholar] [CrossRef] [Green Version]
- Ding, N. Study on Extraction of Passion Fruit Peel Pectin, Derivatization and Functionality. Master Thesis, University of Shanghai for Science & Technology, Shanghai, China, 2020. [Google Scholar]
- Li, X.; Li, S.; Liang, X.; McClements, D.J.; Liu, X.; Liu, F. Applications of oxidases in modification of food molecules and colloidal systems: Laccase, peroxidase and tyrosinase. Trends Food Sci. Tech. 2020, 103, 78–93. [Google Scholar] [CrossRef]
- Vuillemin, M.E.; Muniglia, L.; Linder, M.; Bouguet-Bonnet, S.; Poinsignon, S.; Morais, R.D.S.; Simard, B.; Paris, C.; Michaux, F.; Jasniewski, J. Polymer functionalization through an enzymatic process: Intermediate products characterization and their grafting onto gum Arabic. Int. J. Biol. Macromol. 2021, 169, 480–491. [Google Scholar] [CrossRef]
- Moreno-Expósito, L.; Illescas-Montes, R.; Melguizo-Rodríguez, L.; Ruiz, C.; Ramos-Torrecillas, J.; de Luna-Bertos, E. Multifunctional capacity and therapeutic potential of lactoferrin. Life Sci. 2018, 195, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, X.; Yu, Z.; Wu, S.; Ding, L.; Liu, J. Identification of lactoferrin-derived peptides as potential inhibitors against the main protease of SARS-CoV-2. LWT 2022, 154, 112684. [Google Scholar] [CrossRef] [PubMed]
- Schryvers, A.B. Targeting bacterial transferrin and lactoferrin receptors for vaccines. Trends Microbiol. 2022, 30, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Blumenkrantz, N.; Hansen, G.A. New method for quantitative determination of uronic acids. Anal. Biochem. 1973, 54, 484–489. [Google Scholar] [CrossRef]
- Sato, M.; Ramarathnam, N.; Suzuki, Y.; Ohkubo, T.; Takeuchi, M.; Ochi, H. Varietal differences in the phenolic content and superoxide radical scavenging potential of wines from different sources. J. Agr. Food Chem. 1996, 44, 37–41. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Balance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food–an international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Parameter | Unit | PP | PF Conjugate |
|---|---|---|---|
| Total uronic acid content | (% w/w) | 72.72 ± 2.53 a,2 | 63.19 ± 1.59 b |
| Total phenolic content (TPC) | (mg GAE/g) 1 | 2.36 ± 0.01 b | 20.03 ± 0.38 a |
| Degree of esterification (DE) | (%) | 35.17 ± 1.04 b | 55.40 ± 3.71 a |
| Monosaccharide composition | |||
| Galacturonic acid (GalA) | (mol%) | 53.42 ± 1.23 a | 46.83 ± 0.64 b |
| Arabinose (Ara) | (mol%) | 14.73 ± 0.18 a | 15.16 ± 0.26 a |
| Galactose (Gal) | (mol%) | 18.50 ± 1.28 a | 19.98 ± 1.29 a |
| Glucose (Glc) | (mol%) | 9.11 ± 0.27 b | 16.02 ± 0.50 a |
| Rhamnose (Rha) | (mol%) | 4.24 ± 0.32 a | 2.01 ± 0.06 b |
| HG% = GalA% − Rha% | (mol%) | 49.17 ± 1.46 a | 44.82 ± 0.60 b |
| RG-I% = 2Rha% + Ara% + Gal% | (mol%) | 41.72 ± 1.39 a | 39.16 ± 0.92 a |
| GalA%/(Rha% + Ara% + Gal%) | 1.43 ± 0.08 a | 1.26 ± 0.05 a | |
| Rha%/GalA% | 0.08 ± 0.01 a | 0.04 ± 0.00 b | |
| (Ara% + Gal%)/Rha% | 7.87 ± 0.59 b | 17.53 ± 1.07 a | |
| Targeted Peptide Fragment | Peptide Fragment | Sequence | Sample | |||
|---|---|---|---|---|---|---|
| LfG | LfPFG | LfPFTGG | GI Digests | |||
| DGGMVFEAGRDPYKLRPVAAE, f(79–99) | VFEAGRDPYKLRPVAA | 83–98 | + | + | + | |
| VFEAGRDPYKLRPVA | 83–97 | + | + | + | ||
| FEAGRDPYKLRPVAAE | 84–99 | + | + | + | ||
| FEAGRDPYKLRPVAA | 84–98 | + | + | + | ||
| FEAGRDPYKLRPVA | 84–97 | + | + | + | ||
| FEAGRDPYKLRPV | 84–96 | + | + | + | ||
| EAGRDPYKLRPVAAE | 85–99 | + | + | + | ||
| EAGRDPYKLRPVAA | 85–98 | + | + | + | ||
| EAGRDPYKLRPVA | 85–97 | + | + | + | LfGI (+) | |
| EAGRDPYKLRPV | 85–96 | + | + | + | ||
| AGRDPYKLRPVAAE | 86–99 | + | + | + | ||
| AGRDPYKLRPVAA | 86–98 | + | + | + | ||
| AGRDPYKLRPVA | 86–97 | + | + | + | LfGI (+) | |
| GRDPYKLRPVAA | 87–98 | + | + | + | ||
| GRDPYKLRPVA | 87–97 | + | + | + | ||
| RDPYKLRPVA | 88–97 | + | + | + | ||
| PYKLRPVA | 90–97 | + | + | + | ||
| YKLRPVA | 91–97 | + | + | + | ||
| AGRDPYKLRPV | 86–96 | + | + | |||
| PYKLRPVAAE | 90–99 | + | + | |||
| PYKLRPVAA | 90–98 | + | + | |||
| VFEAGRDPYKLRPV | 83–96 | + | + | |||
| EAGRDPYKLRP | 85–95 | + | + | |||
| DGGMVFEAGRDPYKLRPVA | 79–97 | + | ||||
| VFEAGRDPYKLRPVAAE | 83–99 | + | ||||
| GRDPYKLRPV | 87–96 | + | ||||
| FEAGRDPYKLRP | 84–95 | + | ||||
| DPYKLRPVA | 89–97 | + | ||||
| GILRPYLSWTE, f(149–159) | GILRPYL | 149–155 | + | + | + | |
| ILRPYLSW | 150–157 | + | + | + | ||
| ILRPYL | 150–155 | + | + | + | ||
| RPYLSWT | 152–158 | + | + | + | ||
| RPYLSWTE | 152–159 | + | + | |||
| RPYLSW | 152–157 | + | + | |||
| LRPYLSWT | 151–158 | + | ||||
| LRPYLSW | 151–157 | + | ||||
| GILRPYLSW | 149–157 | + | ||||
| FENLPEKADRDQYEL, f(234–248) | FENLPEKADRDQYE | 234–247 | + | + | + | LfPFTGGI (+) |
| FENLPEKADRDQ | 234–245 | + | + | + | ||
| ENLPEKADRDQYEL | 235–248 | + | + | + | ||
| ENLPEKADRDQYE | 235–247 | + | + | + | ||
| ENLPEKADRDQY | 235–246 | + | + | + | LfPFTGGI (+) | |
| FENLPEKADRDQY | 234–246 | + | + | |||
| ARSVDGKEDLIWKLLSK, f(276–292) | ARSVDGKEDLIWKL | 276–289 | + | + | + | |
| RSVDGKEDLIWKL | 277–289 | + | + | + | ||
| RSVDGKEDLIWKLLSK | 277–292 | + | ||||
| SVDGKEDLIWKLLSK | 278–292 | + | ||||
| YLGSRYLT, f(338–345) | YLGSRYLT | 338–345 | + | + | ||
| YLGSRYL | 338–344 | + | ||||
| VLRPTEGYL, f(445–453) | VLRPTEGYL | 445–453 | + | + | + | |
| VLRPTEGY | 445–452 | + | + | |||
| LFKSETKNLL, f(650–659) | FKSETKNLL | 651–659 | + | + | + | |
| LFKSETKNLL | 650–659 | + | ||||
| Source | Digestion Conditions | Active Peptides | Reference |
|---|---|---|---|
| Bovine Lf | Human gastric juice (HGJ, pH 2.5); human duodenal juice (HDJ, pH 7.0) | Antimicrobial peptide: bLfampin) f(268–288); no bLFcin, f(17–41), detected; 70% peptides from N terminus | [5] |
| Bovine Lf | Porcine pepsin (2540 U/mg, pH 2.5, 37 °C, 4 h) | Antihypertensive (ACE-inhibitory) peptides: LIWKL; LFH; RPYL; LNNSRAP | [9] |
| Kluyveromyces marxianusm Lf | Pepsin (0.02 mg/mL, pH 2.0, 37 °C, 90 min) | Antihypertensive (ACE-inhibitory) peptides: LRP, DPYKLRP, PYKLRP, YKLRP, KLRP, GILRP | [29] |
| Bovine Lf | Porcine pepsin (1:100 w/w), pH 2.0, 37 °C, 90 min; porcine pancreatin (E/S = 1%), pH 7.4, 3 h | ACE inhibitor, anticoagulant peptide: ENLPEKADRD | [6] |
| Bovine Lf | Porcine trypsin (3 U/mg, pH 8.0, 37 °C, 2 h) | Osteoblast-promoting peptide: ENLPEKADRDQYEL | [7] |
| Bovine Lf | Porcine pepsin (3%, 2500 U/mg, pH 2.5, 37 °C, 4 h) | Osteoblast-promoting peptide: FKSETKNLL | [8] |
| Bovine Lf | Porcine pepsin (2000 U/mL, pH 3.0, 37 °C, 2 h) | DGGMVFEAGRDPYKLRPVAAE, GILRPYLSWTE, VLRPTEGYL, YLGSRYLT, ARSVDGKEDLIWKLLSK, FKSETKNLL, ENLPEKADRDQYEL | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, M.; Ji, Y.; Ai, L.; Xie, F.; Wu, Y.; Lai, P.F.H. Improving Effects of Laccase-Mediated Pectin–Ferulic Acid Conjugate and Transglutaminase on Active Peptide Production in Bovine Lactoferrin Digests. Catalysts 2023, 13, 521. https://doi.org/10.3390/catal13030521
Xing M, Ji Y, Ai L, Xie F, Wu Y, Lai PFH. Improving Effects of Laccase-Mediated Pectin–Ferulic Acid Conjugate and Transglutaminase on Active Peptide Production in Bovine Lactoferrin Digests. Catalysts. 2023; 13(3):521. https://doi.org/10.3390/catal13030521
Chicago/Turabian StyleXing, Mingxia, Ying Ji, Lianzhong Ai, Fan Xie, Yan Wu, and Phoency F. H. Lai. 2023. "Improving Effects of Laccase-Mediated Pectin–Ferulic Acid Conjugate and Transglutaminase on Active Peptide Production in Bovine Lactoferrin Digests" Catalysts 13, no. 3: 521. https://doi.org/10.3390/catal13030521
APA StyleXing, M., Ji, Y., Ai, L., Xie, F., Wu, Y., & Lai, P. F. H. (2023). Improving Effects of Laccase-Mediated Pectin–Ferulic Acid Conjugate and Transglutaminase on Active Peptide Production in Bovine Lactoferrin Digests. Catalysts, 13(3), 521. https://doi.org/10.3390/catal13030521