Investigation of the Effect of Plasma Polymerized Siloxane Coating for Enzyme Immobilization and Microfluidic Device Conception
Abstract
:1. Introduction
2. Results and Discussion
2.1. Stability of the Immobilized β-Gal
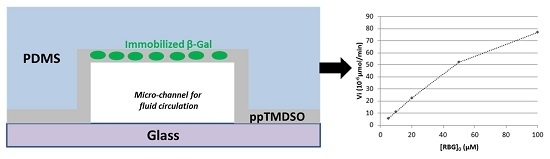
2.2. Kinetic Parameters of Immobilized β-Gal
2.3. Modeling of Physical Phenomena inside the Microsystem
2.4. Diffusion inside the Microsystem
3. Experimental Section and Methods
3.1. Chemicals
3.2. Plasma Polymer Deposition
3.3. Enzyme Immobilization by Entrapment
3.4. Conception of Microreactor with Immobilized Enzyme
3.5. Enzymatic Kinetic Assays
3.6. Modeling of Diffusional Phenomena
4. Conclusions
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Hadd, A.G.; Raymond, D.E.; Halliwell, J.W.; Jacobson, S.C.; Ramsey, J.M. Microchip device for performing enzyme assays. Anal. Chem. 1997, 69, 3407–3412. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Kim, Y.T.; Byun, J.Y.; Ahn, J.; Chung, S.; Gweon, D.G.; Kim, M.G.; Seo, T.S. An integrated allele-specific polymerase chain reaction-microarray chip for multiplex single nucleotide polymorphism typing. Lab Chip 2012, 12, 5146–5154. [Google Scholar] [CrossRef] [PubMed]
- Sassolas, A.; Blum, L.J.; Bouvier, B.D.L. Immobilization strategies to develop enzymatic biosensors. Biotechnol. Adv. 2012, 30, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.W.; Tayyaba, S.; Afzulpurkar, N. Micro Electromechanical Systems (MEMS) based microfluidic devices for biomedical applications. Int. J. Mol. Sci. 2011, 12, 3648–3704. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.Y.; Kim, B. Lab-on-a-Chip pathogen sensors for food safety. Sensors 2012, 12, 10713–10741. [Google Scholar] [CrossRef] [PubMed]
- Ogonczyk, D.; Jankowski, P.; Garstecki, P. Functionalization of polycarbonate with proteins; open-tubular enzymatic microreactors. Lab Chip 2012, 12, 2743–2748. [Google Scholar] [CrossRef] [PubMed]
- Amorosi, C.; Mustin, C.; Frache, G.; Bertani, P.; Fahs, A.; Francius, G.; Toniazzo, V.; Ruch, D.; Ball, V.; Averous, L.; et al. Design of flexible free standing plasma polymer-based films as hosts for enzyme immobilization. J. Phys. Chem. C 2012, 116, 21356–21365. [Google Scholar] [CrossRef]
- Heyries, K.A.; Marquette, C.A.; Blum, L.J. Straightforward protein immobilization on Sylgard 184 PDMS microarray surface. Langmuir 2007, 23, 4523–4527. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Dong, L.; Xu, X.; Shen, S. Immobilization of β-galactosidase from Aspergillus Oryzae on Macroporous PloyGMA Newly Prepared. Int. J. Chem. 2010, 2. [Google Scholar] [CrossRef]
- Elagli, A.; Belhacene, K.; Vivien, C.; Dhulster, P.; Froidevaux, R.; Supiot, P. Facile immobilization of enzyme by entrapment using a plasma-deposited organosilicon thin film. J. Mol. Catal. B Enzym. 2014, 110, 77–86. [Google Scholar] [CrossRef]
- Heyse, P.; van Hoeck, A.; Roeffaers, M.B.J.; Raffin, J.P.; Steinbûchel, A.; Stoveken, T.; Lammertyn, J.; Verboven, P.; Jacobs, P.A.; Hofkens, J.; et al. Exploration of atmospheric pressure plasma nanofilm technology for straightforward bio-active coating deposition: Enzymes, plasmas and polymers, an elegant synergy. Plasma Process. Polym. 2011, 8, 965–974. [Google Scholar] [CrossRef]
- Rabe, M.; Verdes, D.; Seeger, S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Solid Sci. 2011, 1662, 87–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasileva, N.; Iotov, V.; Ivanov, Y.; Godjevargova, T.; Kotia, N. Immobilization of -galactosidase on modified polypropilene membranes. Int. J. Biol. Macromol. 2012, 51, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Dong, M.; Lu, M.; Li, Z. Encapsulation of β-galactosidase from Aspergillus oryzae based on “fish-in-net” approach with molecular imprinting technique. J. Mol. Catal. B Enzym. 2010, 63, 75–80. [Google Scholar] [CrossRef]
- Hanefel, U.; Gardossi, L.; Magner, E. Understanding enzyme immobilisation. Chem. Soc. Rev. 2009, 38, 453–468. [Google Scholar] [CrossRef] [PubMed]
- Bocquet, B.; Bourzgui, N.E.; Guhel, Y.; Mille, V.; Vivien, C.; Supiot, P. Design of silicon-PPTMDS bio-MEMS by cold RPECVD. Proc. SPIE Microfluid. 2004, 5345, 118–129. [Google Scholar]
- Mille, V.; Bourzgui, N.E.; Vivien, C.; Supiot, P.; Bocquet, B. ppTMDS as a new polymer technology for high throughput bio-MEMS design. J. Micromech. Microeng. 2008, 18, 125026. [Google Scholar] [CrossRef]
- Abbas, A.; Treizebre, A.; Supiot, P.; Bourzgui, N.E.; Guillochon, D.; Vercaigne-Marko, D.; Bocquet, B. Cold plasma functionalized TeraHertz BioMEMS for enzyme reaction analysis. Biosens. Bioelectron. 2009, 25, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Supiot, P.; Vivien, C.; Blary, K.; Rouessac, V. Organosilicon polymers deposition by PECVD and RPECVD on micropatterned substrates. Chem. Vap. Depos. 2011, 17, 321–326. [Google Scholar] [CrossRef]
- Urrutia, P.; Colinas, B.R.; Fernandez-Arrojo, L.; Ballesteros, A.O.; Wilson, L.; Illanes, A.; Plou, F.J. Detailed analysis of galactooligosaccharides synthesis with β-galactosidase from Aspergillus oryzae. J. Agric. Food Chem. 2013, 61, 1081–1087. [Google Scholar] [CrossRef] [PubMed]
- Gaur, R.; Pant, H.; Jain, R.; Khare, S.K. Galacto-oligosaccharide synthesis by immobilized Aspergillus oryzae β-galactosidase. Food Chem. 2006, 97, 426–430. [Google Scholar] [CrossRef]
- Gülec, H.; Gürdas, S.; Albayrak, N.; Mutlu, M. Immobilization of Aspergillus oryzae β-Galactosidase on low-pressure plasma-modified cellulose acetate membrane using polyethyleneimine for production of galactooligosaccharide. Biotechnol. Bioprocess Eng. 2010, 15, 1006–1015. [Google Scholar] [CrossRef]
- Gürdaş, S.; Güleç, H.A.; Mutlu, M. Immobilization of Aspergillus oryzae β-Galactosidase onto Duolite A568 Resin via Simple Adsorption Mechanism. Food Bioprocess Technol. 2012, 5, 904–911. [Google Scholar] [CrossRef]
- Mariotti, M.P.; Yamanaka, H.; Araujo, A.R.; Trevisan, H.C. Hydrolysis of whey lactose by immobilized β-galactosidase. Braz. Arch Biol. Technol. 2008, 51, 1233–1240. [Google Scholar] [CrossRef]
- Haubert, K.; Drier, T.; Beebe, D. PDMS bonding by means of a portable, low-cost corona system. Lab Chip 2006, 6, 1548–1549. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Sun, Q.; Wadsworth, L.C. Mechanism of corona treatment on polyolefin films. Polym. Eng. Sci. 1998, 38, 965–970. [Google Scholar] [CrossRef]
- Backer, R.W. Membrane Technology and Applications, 2nd ed.; John Wiley & Son Ltd.: Hoboken, NJ, USA, 2004. [Google Scholar]
- Seong, G.H.; Heo, J.; Crooks, R.M. Measurement of enzyme kinetics using a continuous-flow microfluidic system. Anal. Chem. 2003, 75, 3161–3167. [Google Scholar] [CrossRef] [PubMed]
- Bayramoglu, G.; Tunali, Y.; Arica, M.Y. Immobilization of β-galactosidase onto magnetic poly(GMA–MMA) beads for hydrolysis of lactose in bed reactor. Catal. Commun. 2007, 8, 1094–1101. [Google Scholar] [CrossRef]
- Batchelor, G.K. An Introduction to Fluid Dynamics; Cambridge University Press: Cambridge, UK, 1967; pp. 211–215. [Google Scholar]
- Miller, C.C. The Stokes-Einstein Law for diffusion in solution. Proc. R. Soc. Lond. Ser. A 1924, 106, 724–749. [Google Scholar] [CrossRef]
- Schilling, E.A.; Kamholz, A.E.; Yager, P. Optical measurement of transverse molecular diffusion in a microchannel. Anal. Chem. 2002, 74, 1798–1804. [Google Scholar] [CrossRef] [PubMed]
- Segel, I.H. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems; Wiley Intersciences: New York, NY, USA, 1975; p. 957. [Google Scholar]
- Colowick, S.P.; Kaplan, N.O. Methods in Enzymology. Immobilized Enzymes and Cells (Part C); Academic Press: San Diego, CA, USA, 1987; Volume 136. [Google Scholar]
- Pallares, J.; Ferré, J.A. A simple model to predict mass transfer rates and kinetics of biochemical and biomedical Michaelis-Menten surface reactions. Int. J. Heat Mass Transf. 2015, 80, 192–198. [Google Scholar] [CrossRef]
- Callebert, F.; Supiot, P.; Asfardjani, K.; Dessaut, O.; Gourmaud, P.; Dhamelicourt, P.; Laureyns, J. Cold remote nitrogen plasma polymerization from 1.1.3.3-tetramethyldisiloxane–oxygen mixture. J. Appl. Polym. Sci. 1994, 52, 1595–1606. [Google Scholar] [CrossRef]
- Jambovane, S.; Duin, E.C.; Kim, S.-K.; Hong, J.W. Determination of kinetic parameters, Km and kcat, with a single experiment on a chip. Anal. Chem. 2009, 81, 3239–3245. [Google Scholar] [CrossRef] [PubMed]
- Toepke, M.W.; Beebe, D. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Ghali, N.E.; Vivien, C.; Mutel, B.; Rives, A. Multilayer coating by plasma polymerization of TMDSO deposited on carbon steel: Synthesis and characterization. Surf. Coat. Technol. 2014, 259, 504–516. [Google Scholar] [CrossRef]
- Rich, S.A.; Mille, V.; Vivien, C.; Godey, S.; Supiot, P. Kinetics of RPECVD Organosilicon Polymer Post-treatment in a N2/O2 Microwave Plasma Remote Afterglow. Plasma Process. Polym. 2010, 7, 775–784. [Google Scholar] [CrossRef]








| Catalytic Constants | Km (µM) | Vmax (10−3 µmol·min−1) | Vmaxspé (µmol·min−1·mg−2) | Kcat (s−1) | Kcat /Km (µM−1·s−1) |
|---|---|---|---|---|---|
| Free β-galactosidase | 453 ± 130 | 52 ± 11 | 5.2 | 9.0 | 0.022 |
| Immobilized β-galactosidase | 511 ± 112 | 0.57 ± 0.07 | 0.6 | 1.0 | 0.002 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belhacene, K.; Elagli, A.; Vivien, C.; Treizebré, A.; Dhulster, P.; Supiot, P.; Froidevaux, R. Investigation of the Effect of Plasma Polymerized Siloxane Coating for Enzyme Immobilization and Microfluidic Device Conception. Catalysts 2016, 6, 209. https://doi.org/10.3390/catal6120209
Belhacene K, Elagli A, Vivien C, Treizebré A, Dhulster P, Supiot P, Froidevaux R. Investigation of the Effect of Plasma Polymerized Siloxane Coating for Enzyme Immobilization and Microfluidic Device Conception. Catalysts. 2016; 6(12):209. https://doi.org/10.3390/catal6120209
Chicago/Turabian StyleBelhacene, Kalim, Adil Elagli, Céline Vivien, Anthony Treizebré, Pascal Dhulster, Philippe Supiot, and Renato Froidevaux. 2016. "Investigation of the Effect of Plasma Polymerized Siloxane Coating for Enzyme Immobilization and Microfluidic Device Conception" Catalysts 6, no. 12: 209. https://doi.org/10.3390/catal6120209
APA StyleBelhacene, K., Elagli, A., Vivien, C., Treizebré, A., Dhulster, P., Supiot, P., & Froidevaux, R. (2016). Investigation of the Effect of Plasma Polymerized Siloxane Coating for Enzyme Immobilization and Microfluidic Device Conception. Catalysts, 6(12), 209. https://doi.org/10.3390/catal6120209