Efficient Conversion of Acetate to 3-Hydroxypropionic Acid by Engineered Escherichia coli
Abstract
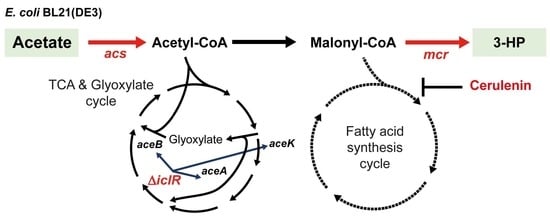
:1. Introduction
2. Results
2.1. Heterologous Expression of mcr for 3-HP Production from Acetate
2.2. Engineering the Acetate Assimilation and Glyoxylate Shunt Pathways
2.3. Improved 3-HP Production from Acetate by Inhibiting Fatty Acid Synthesis
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Plasmid Cloning and Bacterial Strain Construction
4.3. Cultivation Methods
4.4. Analytical Methods
4.5. Calculation of the Theoretical Maximum Yield
Author Contributions
Funding
Conflicts of Interest
References
- Kidanu, W.G.; Trang, P.T.; Yoon, H.H. Hydrogen and volatile fatty acids production from marine macroalgae by anaerobic fermentation. Biotechnol. Bioprocess Eng. 2017, 22, 612–619. [Google Scholar] [CrossRef]
- Shrestha, B.; Dhakal, D.; Darsandhari, S.; Pandey, R.P.; Pokhrel, A.R.; Jnawali, H.N.; Sohng, J.K. Heterologous production of clavulanic acid intermediates in Streptomyces venezuelae. Biotechnol. Bioprocess Eng. 2017, 22, 359–365. [Google Scholar] [CrossRef]
- Luo, X.; Ge, X.; Cui, S.; Li, Y. Value-added processing of crude glycerol into chemicals and polymers. Bioresour. Technol. 2016, 215, 144–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, N.; Quarterman, J.; Jin, Y.-S. Marine macroalgae: An untapped resource for producing fuels and chemicals. Trends Biotechnol. 2013, 31, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.-J.; Galazka, J.M.; Kim, S.R.; Choi, J.-H.; Yang, X.; Seo, J.-H.; Glass, N.L.; Cate, J.H.D.; Jin, Y.-S. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc. Natl. Acad. Sci. USA 2011, 108, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Yang, H.; Fang, G.; Zhang, X.; Wu, H.; Li, Z.; Ye, Q. Central pathway engineering for enhanced succinate biosynthesis from acetate in Escherichia coli. Biotechnol. Bioeng. 2018, 115, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Novak, K.; Pflügl, S. Towards biobased industry: Acetate as a promising feedstock to enhance the potential of microbial cell factories. FEMS Microbiol. Lett. 2018, 365, fny226. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.G.; Lee, J.H.; Noh, M.H.; Jung, G.Y. Rediscovering Acetate Metabolism: Its Potential Sources and Utilization for Biobased Transformation into Value-Added Chemicals. J. Agric. Food Chem. 2018, 66, 3998–4006. [Google Scholar] [CrossRef] [PubMed]
- Leone, S.; Sannino, F.; Tutino, M.L.; Parrilli, E.; Picone, D. Acetate: Friend or foe? Efficient production of a sweet protein in Escherichia coli BL21 using acetate as a carbon source. Microb. Cell Fact. 2015, 14, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Nie, Q. Engineering Escherichia coli to convert acetic acid to β-caryophyllene. Microb. Cell Fact. 2016, 15, 74. [Google Scholar] [CrossRef] [PubMed]
- Noh, M.H.; Lim, H.G.; Woo, S.H.; Song, J.; Jung, G.Y. Production of itaconic acid from acetate by engineering acid-tolerant Escherichia coli W. Biotechnol. Bioeng. 2018, 115, 729–738. [Google Scholar] [CrossRef] [PubMed]
- de Fouchécour, F.; Sánchez-Castañeda, A.-K.; Saulou-Bérion, C.; Spinnler, H.É. Process engineering for microbial production of 3-hydroxypropionic acid. Biotechnol. Adv. 2018, 36, 1207–1222. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Ashok, S.; Park, S. Recent advances in biological production of 3-hydroxypropionic acid. Biotechnol. Adv. 2013, 31, 945–961. [Google Scholar] [CrossRef] [PubMed]
- Seok, J.Y.; Yang, J.; Choi, S.J.; Lim, H.G.; Choi, U.J.; Kim, K.-J.; Park, S.; Yoo, T.H.; Jung, G.Y. Directed evolution of the 3-hydroxypropionic acid production pathway by engineering aldehyde dehydrogenase using a synthetic selection device. Metab. Eng. 2018, 47, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.G.; Noh, M.H.; Jeong, J.H.; Park, S.; Jung, G.Y. Optimum Rebalancing of the 3-Hydroxypropionic Acid Production Pathway from Glycerol in Escherichia coli. ACS Synth. Biol. 2016, 5, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ding, Y.; Zhang, R.; Liu, H.; Xian, M.; Zhao, G. Functional balance between enzymes in malonyl-CoA pathway for 3-hydroxypropionate biosynthesis. Metab. Eng. 2016, 34, 104–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rathnasingh, C.; Raj, S.M.; Lee, Y.; Catherine, C.; Ashok, S.; Park, S. Production of 3-hydroxypropionic acid via malonyl-CoA pathway using recombinant Escherichia coli strains. J. Biotechnol. 2012, 157, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Jiang, J.; Wu, H.; Li, Z.; Ye, Q. Enhanced production of 3-hydroxypropionic acid from glucose via malonyl-CoA pathway by engineered Escherichia coli. Bioresour. Technol. 2016, 200, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ding, Y.; Xian, M.; Liu, M.; Liu, H.; Ma, Q.; Zhao, G. Malonyl-CoA pathway: A promising route for 3-hydroxypropionate biosynthesis. Crit. Rev. Biotechnol. 2017, 37, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; Millard, P.; Dinclaux, M.; Portais, J.-C.; Létisse, F. Acetate fluxes in Escherichia coli are determined by the thermodynamic control of the Pta-AckA pathway. Sci. Rep. 2017, 7, 42135. [Google Scholar] [CrossRef] [PubMed]
- Rajaraman, E.; Agarwal, A.; Crigler, J.; Seipelt-Thiemann, R.; Altman, E.; Eiteman, M.A. Transcriptional analysis and adaptive evolution of Escherichia coli strains growing on acetate. Appl. Microbiol. Biotechnol. 2016, 100, 7777–7785. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.; Yeow, J.; Wang, I.; Song, H.; Jiang, R. Improving acetate tolerance of Escherichia coli by rewiring its global regulator cAMP receptor protein (CRP). PLoS ONE 2013, 8, e77422. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sandoval, M.T.; Huerta-Beristain, G.; Trujillo-Martinez, B.; Bustos, P.; González, V.; Bolivar, F.; Gosset, G.; Martinez, A. Laboratory metabolic evolution improves acetate tolerance and growth on acetate of ethanologenic Escherichia coli under non-aerated conditions in glucose-mineral medium. Appl. Microbiol. Biotechnol. 2012, 96, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, C.; Maurer, L.M.; Oyelakin, N.E.; Yoncheva, Y.N.; Maurer, R.; Slonczewski, J.L. Acetate and formate stress: Opposite responses in the proteome of Escherichia coli. J. Bacteriol. 2001, 183, 6466–6477. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Vora, H.; Khosla, C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab. Eng. 2010, 12, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lin, Y.; Li, L.; Linhardt, R.J.; Yan, Y. Regulating malonyl-CoA metabolism via synthetic antisense RNAs for enhanced biosynthesis of natural products. Metab. Eng. 2015, 29, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Zha, W.; Rubin-Pitel, S.B.; Shao, Z.; Zhao, H. Improving cellular malonyl-CoA level in Escherichia coli via metabolic engineering. Metab. Eng. 2009, 11, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.W.; Yang, J.-S.; Kim, I.; Yang, J.; Min, B.E.; Kim, S.; Jung, G.Y. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 2013, 15, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Noh, M.H.; Lim, H.G.; Park, S.; Seo, S.W.; Jung, G.Y. Precise flux redistribution to glyoxylate cycle for 5-aminolevulinic acid production in Escherichia coli. Metab. Eng. 2017, 43, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Holms, W.H.; Bennett, P.M. Regulation of isocitrate dehydrogenase activity in Escherichia coli on adaptation to acetate. J. Gen. Microbiol. 1971, 65, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Moche, M.; Schneider, G.; Edwards, P.; Dehesh, K.; Lindqvist, Y. Structure of the complex between the antibiotic cerulenin and its target, beta-ketoacyl-acyl carrier protein synthase. J. Biol. Chem. 1999, 274, 6031–6034. [Google Scholar] [CrossRef] [PubMed]
- Price, A.C.; Choi, K.H.; Heath, R.J.; Li, Z.; White, S.W.; Rock, C.O. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism. J. Biol. Chem. 2001, 276, 6551–6559. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.K.; Church, G.M. Genetically encoded sensors enable real-time observation of metabolite production. Proc. Natl. Acad. Sci. USA 2016, 113, 2388–2393. [Google Scholar] [CrossRef] [PubMed]
- Menzella, H.G. Comparison of two codon optimization strategies to enhance recombinant protein production in Escherichia coli. Microb. Cell Fact. 2011, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Anthony, L.C.; Nowroozi, F.; Kwon, G.; Newman, J.D.; Keasling, J.D. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab. Eng. 2009, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Yim, H.; Haselbeck, R.; Niu, W.; Pujol-Baxley, C.; Burgard, A.; Boldt, J.; Khandurina, J.; Trawick, J.D.; Osterhout, R.E.; Stephen, R.; et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011, 7, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]




| Gene | 5′ UTR sequence (5′−3′) a | Predicted Expression Level (a.u.) |
|---|---|---|
| mcr | AACAATTACTAGTAAGGAGAGGAGT | 3,110,669.92 |
| acs | AAAATCAGCGCCCAAGGAGTCACCG b | 1,074,836.02 |
| Name | Description | Source |
|---|---|---|
| Strains | ||
| Mach1-T1R | E. coli F− φ80(lacZ)ΔM15 ΔlacX74 hsdR(rK−mK+) ΔrecA1398 endA1 tonA | Invitrogen |
| BL21(DE3) | E. coli F− ompT gal dcm lon hsdSB (rB− mB−) λ(DE3) | Invitrogen |
| HJ1 | BL21(DE3)/pET-mcr* | This study |
| HJ2 | BL21(DE3)/pET-mcr*/pACYC-acs | This study |
| HJ3 | BL21(DE3) ΔiclR/pET-mcr* | This study |
| HJ4 | BL21(DE3) ΔiclR/pET-mcr*/pACYC-acs | This study |
| Plasmids | ||
| PLB0110 | Source of mcr | [15] |
| pETDuet | Expression vector, ColE1 ori, AmpR | Novagen |
| pACYCDuet | Expression vector, p15A ori, CmR | Novagen |
| pKD46 | Red recombinase expression vector, AmpR | [32] |
| pFRT72variant | Source of mutant FRT-kanR-FRT | [33] |
| pCP20 | FLP expression vector, AmpR, CmR | [32] |
| pET-mcr* | pETDuet/Ptac-SynUTRmcr-mcr N940V, K1106W, S1114R | This study |
| pACYC-acs | pACYCDuet/PBBa_J23100-SynUTRacs-acs | This study |
| Name | Sequence (5′−3′) | |
|---|---|---|
| O-mcr-F1 | GGAATTGTGAGCGGATAACAATTACTAGTAAGGAGAGGAGTATGAGCGGAACAGGACGACT | |
| O-mcr-F2 | GGATCCTTGACAATTAATCATCGGCTCGTATAATGTGTGGAATTGTGAGCGGATAACAATT | |
| O-mcr-B | CTCGAGTGCGAAAAAACCCCGCCGAAGCGGGGTTTTTTGCGGCATGCTTACACGGTAATCGCCCGT | |
| O-N940V-F | TATTACCTTGCCGACCGCAATGTCAGTGGTGAGACATTCC | |
| O-N940V-B | GCGGTCGGCAAGGTAATAG | |
| O-K1106W-F | ATTTCCGGGTAGCGCGCAAGATTGCCCTGAGTGATGGTG | |
| O-K1106W-B | GCGCGCTACCCGGAAATG | |
| O-S1114R-F | TGAGTGATGGTGCCAGTCTCGCGCTGGTCACTC | |
| O-S1114R-B | AGACTGGCACCATCACTCAGGGC | |
| O-acs-F | GAATTCTTGACGGCTAGCTCAGTCCTAGGTACAGTGCTAGCAAAATCAGCGCCCAAGGAGTCACCGATGAGCCAAATTCACAAACACA | |
| O-acs-B | GAGCTCAAAAAAAACCCCGCCCTGTCAGGGGCGGGGTTTTTTTTTTTACGATGGCATCGCGATAG | |
| R-iclR-F | TGCCACTCAGGTATGATGGGCAGAATATTGCCTCTGCCCGCCAGAAAAAGGCATGACCGGCGCGATGC | |
| R-iclR-B | TAACAATAAAAATGAAAATGATTTCCACGATACAGAAAAAGGAGACTGTCGCTCAGCGGATCTCATGCGC | |
| C-iclR-F | CAACATTAACTCATCGGATCAG | |
| C-iclR-B | TCTATTGCCACTCAGGTATGATGGGC | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.H.; Cha, S.; Kang, C.W.; Lee, G.M.; Lim, H.G.; Jung, G.Y. Efficient Conversion of Acetate to 3-Hydroxypropionic Acid by Engineered Escherichia coli. Catalysts 2018, 8, 525. https://doi.org/10.3390/catal8110525
Lee JH, Cha S, Kang CW, Lee GM, Lim HG, Jung GY. Efficient Conversion of Acetate to 3-Hydroxypropionic Acid by Engineered Escherichia coli. Catalysts. 2018; 8(11):525. https://doi.org/10.3390/catal8110525
Chicago/Turabian StyleLee, Ji Hoon, Sanghak Cha, Chae Won Kang, Geon Min Lee, Hyun Gyu Lim, and Gyoo Yeol Jung. 2018. "Efficient Conversion of Acetate to 3-Hydroxypropionic Acid by Engineered Escherichia coli" Catalysts 8, no. 11: 525. https://doi.org/10.3390/catal8110525
APA StyleLee, J. H., Cha, S., Kang, C. W., Lee, G. M., Lim, H. G., & Jung, G. Y. (2018). Efficient Conversion of Acetate to 3-Hydroxypropionic Acid by Engineered Escherichia coli. Catalysts, 8(11), 525. https://doi.org/10.3390/catal8110525