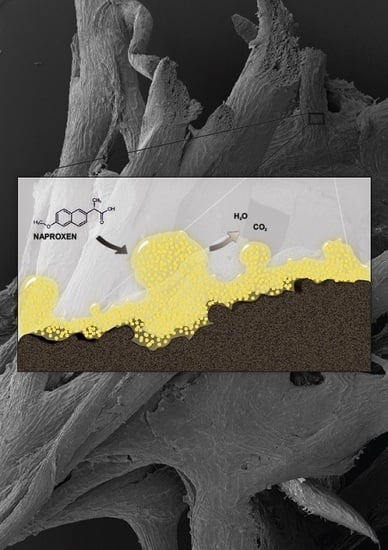
Immobilization of Planococcus sp. S5 Strain on the Loofah Sponge and Its Application in Naproxen Removal
Abstract
:1. Introduction
2. Results and Discussion
2.1. Immobilization of Planococcus sp. S5 on Loofah Sponge
2.2. Naproxen Biodegradation
2.2.1. Biodegradation of Different Concentration of Naproxen
2.2.2. Stability and Degradation Capacity of the Developed Whole-Cell Biocatalyst System
2.3. The Influence of Immobilization on Enzymes Activity
2.4. Changes in Biofilm Formed Onto the Loofah Sponge during Naproxen Degradation
3. Materials and Methods
3.1. Bacterial Cultures Cultivation
3.2. Carrier Preparation for Immobilization
3.3. Immobilization Procedure
3.4. Characterization of Immobilized Loofah Sponges
3.5. Biodegradation Experiments
3.6. Determination of Naproxen Concentration
3.7. Enzyme Assay
3.8. Scanning Electron Microscopy
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Wojcieszyńska, D.; Domaradzka, D.; Hupert-Kocurek, K.; Guzik, U. Enzymes Involved in Naproxen Degradation by Planococcus sp. S5. Pol. J. Microbiol. 2016, 1, 177–182. [Google Scholar] [CrossRef]
- Domaradzka, D.; Guzik, U.; Hupert-Kocurek, K.; Wojcieszyńska, D. Cometabolic degradation of naproxen by Planococcus sp. strain S5. Water Air Soil Pollut. 2015, 226, 297. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Patrolecco, L.; Ademollo, N.; Tolomei, A.; Caracciolo, A.B. Degradation of gemfibrozil and naproxen in a river water ecosystem. Microchem. J. 2013, 107, 158–164. [Google Scholar] [CrossRef]
- Marotta, R.; Spasiano, D.; Di Somma, I.; Andreozzi, R. Photodegradation of naproxen and its photoproducts in aqueous solution at 254 nm: A kinetic investigation. Water Res. 2013, 47, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musa, K.A.; Eriksson, L.A. Theoretical study of the phototoxicity of naproxen and the active form of nabumetone. J. Phys. Chem. A 2008, 112, 10921–10930. [Google Scholar] [CrossRef] [PubMed]
- Isidori, M.; Lavorgna, M.; Nardelli, A.; Parrella, A.; Previtera, L.; Rubino, M. Ecotoxicity of naproxen and its phototransformation products. Sci. Total Environ. 2005, 348, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.; Saez, J.M.; Costa, J.S.D.; Colin, V.L.; Fuentes, M.S.; Cuozzo, S.A.; Amoroso, M.J. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Dzionek, A.; Wojcieszyńska, D.; Guzik, U. Natural carriers in bioremediation: A review. Electron. J. Biotechnol. 2016, 19, 28–36. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, M. Bioremediation of crude oil-contaminated soil: Comparison of different biostimulation and bioaugmentation treatments. J. Hazard. Mater. 2010, 183, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Alessandrello, M.J.; Parellada, E.A.; Tomás, M.S.J.; Neske, A.; Vullo, D.L.; Ferrero, M.A. Polycyclic aromatic hydrocarbons removal by immobilized bacterial cells using annonaceous acetogenins for biofilm formation stimulation on polyurethane foam. J. Environ. Chem. Eng. 2017, 5, 189–195. [Google Scholar] [CrossRef]
- Carpentier, B.; Cerf, O. Biofilms and their consequences, with particular reference to hygiene in the food industry. J. Appl. Microbiol. 1993, 75, 499–511. [Google Scholar] [CrossRef]
- Bayat, Z.; Hassanshahian, M.; Cappello, S. Immobilization of microbes for bioremediation of crude oil polluted environments: A mini review. Open Microbiol. J. 2015, 9, 48–54. [Google Scholar] [PubMed]
- Moreno-Medina, D.A.; Sánchez-Salinas, E.; Ortiz-Hernández, M.L. Removal of methyl parathion and coumaphos pesticides by a bacterial consortium immobilized in Luffa cylindrica. Rev. Int. Contam. Ambient. 2014, 30, 51–63. [Google Scholar]
- Iqbal, M.; Saeed, A.; Edyvean, R.G.J.; O’Sullivan, B.; Styring, P. Production of fungal biomass immobilized loofa sponge (FBILS)-discs for the removal of heavy metal ions and chlorinated compounds from aqueous solution. Biotechnol. Lett. 2005, 27, 1319–1323. [Google Scholar] [CrossRef] [PubMed]
- Mazmanci, M.A.; Ünyayar, A. Decolourisation of reactive black 5 by Funalia trogii immobilised on Luffa cylindrica sponge. Process Biochem. 2005, 40, 337–342. [Google Scholar] [CrossRef]
- Picioreanu, C.; Van Loosdrecht, M.C.; Heijnen, J.J. Two-dimensional model of biofilm detachment caused by internal stress from liquid flow. Biotechnol. Bioeng. 2001, 72, 205–218. [Google Scholar] [CrossRef]
- Ohashi, A.; Harada, H. Adhesion strength of biofilm developed in an attached-growth reactor. Water Sci. Technol. 1994, 29, 281–288. [Google Scholar]
- Stanley, P.M. Factors affecting the irreversible attachment of Pseudomonas aeruginosa to stainless steel. Can. J. Microbiol. 1983, 29, 1493–1499. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Huang, J.; Lu, H.; Liu, J.; Yan, C. Optimisation for assay of fluorescein diacetate hydrolytic activity as a sensitive tool to evaluate impacts of pollutants and nutrients on microbial activity in coastal sediments. Mar. Pollut. Bull. 2016, 110, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Fontvieille, D.A.; Outaguerouine, A.; Thevenot, D.R. Fluorescein diacetate hydrolysis as a measure of microbial activity in aquatic systems: Application to activated sludges. Environ. Technol. 1992, 13, 531–540. [Google Scholar] [CrossRef]
- Łabużek, S.; Hupert-Kocurek, K.; Skurnik, M. Isolation and characterisation of new Planococcus sp. strain able for aromatic hydrocarbons degradation. Acta Microbiol. Pol. 2003, 52, 395–404. [Google Scholar] [PubMed]
- Hupert-Kocurek, K.; Guzik, U.; Wojcieszynska, D. Characterization of catechol 2,3-dioxygenase from Planococcus sp. strain S5 induced by high phenol concentration. Acta Biochem. Pol. 2012, 59, 345–351. [Google Scholar]
- Qurashi, A.W.; Sabri, A.N. Bacterial exopolysaccharide and biofilm formation stimulate chickpea growth and soil aggregation under salt stress. Braz. J. Microbiol. 2012, 43, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Sonenshein, A.L. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 2000, 3, 561–566. [Google Scholar] [CrossRef]
- Gotz, F. Staphylococcus and biofilms. Mol. Microbiol. 2002, 43, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Y.H.; Luo, N.; Zhang, X.Y.; Luan, T.G.; Hu, J.M.; Wang, Z.Y.; Wu, P.C.; Chen, M.J.; Lu, J.Q. Biodegradation of benzene and its derivatives by a psychrotolerant and moderately haloalkaliphilic Planococcus sp. strain ZD22. Res. Microbiol. 2006, 157, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Romano, I.; Giordano, A.; Lama, L.; Nicolaus, B.; Gambacorta, A. Planococcus rifietensis sp. nov, Isolated from Algal Mat Collected from a Sulfurous Spring in Campania (Italy). Syst. Appl. Microbiol. 2003, 26, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Sprott, G.D.; Larocque, S.; Cadotte, N.; Dicaire, C.J.; McGee, M.; Brisson, J.R. Novel polar lipids of halophilic eubacterium Planococcus H8 and archaeon Haloferax volcanii. Biochim. Biophys. Acta 2003, 1633, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Marco-Urrea, E.; Pérez-Trujillo, M.; Blánquez, P.; Vicent, T.; Caminal, G. Biodegradation of the analgesic naproxen by Trametes versicolor and identification of intermediates using HPLC-DAD-MS and NMR. Bioresour. Technol. 2010, 101, 2159–2166. [Google Scholar] [CrossRef] [PubMed]
- Rodarte-Morales, A.I.; Feijoo, G.; Moreira, M.T.; Lema, J.M. Degradation of selected pharmaceutical and personal care products (PPCPs) by white-rot fungi. World J. Microbiol. Biotechnol. 2011, 27, 1839–1846. [Google Scholar] [CrossRef]
- Marchlewicz, A.; Domaradzka, D.; Guzik, U.; Wojcieszyńska, D. Bacillus thuringiensis B1 (2015b) is a Gram-positive bacteria able to degrade naproxen and ibuprofen. Water Air Soil Pollut. 2016, 227, 197. [Google Scholar] [CrossRef] [PubMed]
- Wojcieszyńska, D.; Domaradzka, D.; Hupert-Kocurek, K.; Guzik, U. Bacterial degradation of naproxen–Undisclosed pollutant in the environment. J. Environ. Manag. 2014, 145, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Lyu, J.; Lyu, X.J.; Yu, H.Q.; Hu, Z.; Lam, J.C.; Lam, P.K. Characterization of cefalexin degradation capabilities of two Pseudomonas strains isolated from activated sludge. J. Hazard. Mater. 2015, 282, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Hu, N.; Wang, Z. Experimental investigation of Luffa cylindrica as a natural sorbent material for the removal of a cationic surfactant. J. Taiwan Inst. Chem. Eng. 2013, 44, 74–80. [Google Scholar] [CrossRef]
- Demir, H.; Top, A.; Balköse, D.; Ülkü, S. Dye adsorption behaviour of Luffa cylindrica fibres. J. Hazard. Mater. 2008, 153, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Huerta, B.; Rodriguez-Mozaz, S.; Nannou, C.; Nakis, L.; Ruhi, A.; Acuña, V.; Barcelo, D. Determination of a broad spectrum of pharmaceuticals and endocrine disruptors in biofilm from a waste water treatment plant-impacted river. Sci. Total Environ. 2016, 540, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Neeraja, P.; Srinivas, S.; Mukkanti, K.; Dubey, P.K.; Pal, S. 1H-1, 2, 3-Triazolyl-substituted 1, 3, 4-oxadiazole derivatives containing structural features of ibuprofen/naproxen: Their synthesis and antibacterial evaluation. Bioorg. Med. Chem. Lett. 2016, 26, 5212–5217. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Wang, Y.; Whittell, L.R.; Jergic, S.; Liu, M.; Harry, E.; Oakley, A.J. DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs. Chem. Biol. 2014, 21, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Gunsch, C.K. Effects of selected pharmaceutically active compounds on the ammonia oxidizing bacterium Nitrosomonas europaea. Chemosphere 2011, 82, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Rahman, R.N.Z.A.; Ghazali, F.M.; Salleh, A.B.; Basri, M. Biodegradation of hydrocarbon contamination by immobilized bacterial cells. J. Microbiol. 2006, 44, 354–359. [Google Scholar] [PubMed]
- Lloret, L.; Eibes, G.; Lú-Chau, T.A.; Moreira, M.T.; Feijoo, G.; Lema, J.M. Laccase-catalysed degradation of anti-inflammatories and estrogens. Biochem. Eng. J. 2010, 51, 124–131. [Google Scholar] [CrossRef]
- Rodríguez-Rodríguez, C.E.; Marco-Urrea, E.; Caminal, G. Naproxen degradation test to monitor Trametes versicolor activity in solid-state bioremediation processes. J. Hazard. Mater. 2010, 179, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.H.; Urase, T.; Kusakabe, O. Biodegradation characteristics of pharmaceutical substances by whole fungal culture Trametes versicolor and its laccase. J. Water Environ. Technol. 2010, 8, 125–140. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszyńska, D. Intradiol dioxygenases—The key enzymes in xenobiotics degradation in biodegradation of hazardous and special products. InTech 2013, 7, 129–153. [Google Scholar]
- Cidaria, D.; Deidda, F.; Bosetti, A. A rapid method for naphtalene dioxygenase assay in whole cells of naphtalene cis-dihydrodiol dehydrogenase blocked Pseudomonas fluoresecens: Screening of potential inducers of dioxygenase activity. Appl. Microbiol. Biotechnol. 1994, 41, 689–693. [Google Scholar] [CrossRef]
- Jõesaar, M.; Viggor, S.; Heinaru, E.; Naanuri, E.; Mehike, M.; Leito, I.; Heinaru, A. Strategy of Pseudomonas pseudoalcaligenes C70 for effective degradation of phenol and salicylate. PLoS ONE 2017, 12, e0173180. [Google Scholar] [CrossRef] [PubMed]
- Hintner, J.P.; Lechner, C.; Riegert, U.; Kuhm, A.E.; Storm, T.; Reemtsma, T.; Stolz, A. Direct ring fission of salicylate by a salicylate 1, 2-dioxygenase activity from Pseudaminobacter salicylatoxidans. J. Bacteriol. 2001, 183, 6936–6942. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Khoo, H.E.; Poh, C.L. Purification and characterization of gentisate 1, 2-dioxygenases from Pseudomonas alcaligenes NCIB 9867 and Pseudomonas putida NCIB 9869. J. Appl. Environ. Microbiol. 1999, 65, 946–950. [Google Scholar]
- Tuson, H.H.; Weibel, D.B. Bacteria–surface interactions. Soft Matter 2013, 9, 4368–4380. [Google Scholar] [CrossRef] [PubMed]
- Mirpuri, R.; Jones, W.; Bryers, J.D. Toluene degradation kinetics for planktonic and biofilm-grown cells of Pseudomonas putida 54G. Biotechnol. Bioeng. 1997, 53, 535–546. [Google Scholar] [CrossRef]
- Priester, J.H.; Horst, A.M.; Van De Werfhorst, L.C.; Saleta, J.L.; Mertes, L.A.; Holden, P.A. Enhanced visualization of microbial biofilms by staining and environmental scanning electron microscopy. J. Microbiol. Methods 2007, 68, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Asahi, Y.; Miura, J.; Tsuda, T.; Kuwabata, S.; Tsunashima, K.; Noiri, Y.; Hayashi, M. Simple observation of Streptococcus mutans biofilm by scanning electron microscopy using ionic liquids. AMB Express 2015, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.D.; Dong, S.S.; Gao, L.L.; Liu, M.Y.; Niu, S. Distribution characteristics of extracellular polymeric substances and cells of aerobic granules cultivated in a continuous-flow airlift reactor. J. Chem. Technol. Biotechnol. 2013, 88, 942–947. [Google Scholar] [CrossRef]
- Ma, D.; Zou, D.; Zhou, D.; Li, T.; Dong, S.; Xu, Z.; Dong, S. Phenol removal and biofilm response in coupling of visible-light-driven photocatalysis and biodegradation: Effect of hydrothermal treatment temperature. Int. Biodeterior. Biodegrad. 2015, 104, 178–185. [Google Scholar] [CrossRef]
- Greń, I.; Wojcieszyńska, D.; Guzik, U.; Perkosz, M.; Hupert-Kocurek, K. Enhanced biotransformation of mononitrophenols by Stenotrophomonas malthophilia KB2 in the presence of aromatic compounds of plant origin. World J. Microbiol. Biotechnol. 2010, 26, 289–295. [Google Scholar] [CrossRef]
- Abe, T.; Masai, E.; Miyauchi, K.; Katayama, Y.; Fukuda, M. A tetrahydrofolate-dependent O-demethylase, LigM, is crucial for catabolism of vanillate and syringate in Sphingomonas paucimobilis SYK-6. J. Bacteriol. 2005, 187, 2030–2037. [Google Scholar] [CrossRef] [PubMed]
- Wojcieszyńska, D.; Greń, I.; Hupert-Kocurek, K.; Guzik, U. Modulation of FAD-dependent monooxygenase activity from aromatic compounds-degrading Stenotrophomonas maltophilia strain KB2. Acta Biochem. Pol. 2011, 58, 421–426. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]






| Period [Day–Day] | Average Naproxen Degradation Rate [µg/h] | |||
|---|---|---|---|---|
| 6 mg/L | 9 mg/L | 12 mg/L | 15 mg/L | |
| 0–3 | 9.8 ± 2.2 | 6.6 ± 0.6 | 6.4 ± 1.1 | 5.7 ± 0.9 |
| 3–7 | 4.8 ± 2.2 | 7.3 ± 1.7 | 5.8 ± 1.0 | 5.0 ± 1.7 |
| 7–11 | 1.3 ± 0.5 | 5.3 ± 2.5 | 1.2 ± 0.5 | 7.6 ± 0.2 |
| 11–14 | 2.6 ± 1.4 | 7.7 ± 1.5 | 1.7 ± 2.6 | 8.1 ± 1.9 |
| 14–21 | 3.0 ± 0.4 | 2.5 ± 0.4 | 6.4 ± 1.5 | 7.4 ± 0.1 |
| 21–27 | 6.5 ± 1.4 | 4.5 ± 1.7 | 6.4 ± 0.5 | 3.8 ± 0.6 |
| 27–29 | 19.6 ± 3.3 | 19.2 ± 6.0 | 11.5 ± 4.6 | 5.5 ± 1.2 |
| 29–34 | 11.9 ± 0.9 | 10.6 ± 1.4 | 9.2 ± 1.5 | 0.1 ± 1.9 |
| 34–38 | 8.2 ± 1.6 | 18.4 ± 1.3 | 19.6 ± 0.6 | 1.0 ± 3.2 |
| 38–41 | 11.4 ± 0.5 | 10.1 ± 0.7 | 0.6 ± 7.3 | |
| 41–44 | 10.3 ± 0.6 | 6.7 ± 1.1 | −2.2 ± 4.9 | |
| 44–47 | 8.9 ± 5.1 | 1.3 ± 3.8 | ||
| 47–50 | 5.5 ± 4.0 | 1.2 ± 4.0 | ||
| 50–54 | 11.4 ± 0.9 | −1.3 ± 3.7 | ||
| 54–58 | 12.8 ± 1.1 | −0.7 ± 2.4 | ||
| 58–62 | 7.8 ± 2.0 | 1.0 ± 1.9 | ||
| Period [Day–Day] | Average Naproxen Degradation Rate [µg/h] | |||
|---|---|---|---|---|
| 6 mg/L | 9 mg/L | 12 mg/L | 15 mg/L | |
| 0–4 | 11.2 ± 1.2 | 20.7 ± 8.6 | 16.1 ± 6.7 | 13.9 ± 0.6 |
| 4–8 | 13.9 ± 0.9 | 10.1 ± 1.4 | 18.0 ± 14.6 | 10.5 ± 2.5 |
| 8–13 | 15.8 ± 1.1 | 12.9 ± 1.8 | 13.2 ± 5.4 | 13.4 ± 4.2 |
| 13–17 | 18.2 ± 1.7 | 6.4 ± 2.4 | 10.1 ± 1.2 | 10.0 ± 1.4 |
| 17–21 | 11.6 ± 3.0 | 9.4 ± 0.9 | 10.0 ± 1.0 | |
| 21–25 | 15.0 ± 5.8 | 12.0 ± 4.8 | 4.6 ± 4.2 | |
| 25–29 | 7.6 ± 3.9 | 5.1 ± 0.6 | 9.7 ± 5.6 | |
| 29–32 | 8.7 ± 4.2 | 6.4 ± 2.9 | 13.1 ± 5.6 | |
| 32–36 | 11.5 ± 1.7 | 10.8 ± 6.0 | ||
| 36–39 | 17.4 ± 4.8 | 9.4 ± 4.3 | ||
| 39–43 | 7.3 ± 2.4 | 13.0 ± 2.5 | ||
| 43–46 | 19.0 ± 2.8 | |||
| 46–49 | 17.7 ± 0.6 | |||
| 49–53 | 14.6 ± 2.2 | |||
| Period [Day–Day] | Average Naproxen Degradation Rate [µg/h] | |||
|---|---|---|---|---|
| I Cycle | II Cycle | III Cycle | IV Cycle | |
| 0–4 | 11.2 ± 1.2 a | |||
| 4–8 | 13.9 ± 0.9 b | |||
| 8–13 | 15.8 ± 1.1 b | |||
| 13–17 | 18.2 ± 1.7 c | |||
| 17–21 | 23.2 ± 1.8 a | |||
| 21–25 | 16.8 ± 0.7 b | |||
| 25–29 | 17.4 ± 0.2 b | |||
| 29–32 | 6.7 ± 0.1 c | |||
| 32–36 | 15.0 ± 3.8 a | |||
| 36–39 | 8.9 ± 1.4 b | |||
| 39–43 | 9.6 ± 1.2 b | |||
| 43–46 | 18.2 ± 2.0 a | |||
| 46–49 | 12.3 ± 2.4 ab | |||
| 49–53 | 8.3 ± 1.8 b | |||
| 53–55 | 3.8 ± 3.5 a | |||
| 55–58 | 8.5 ± 3.2 b | |||
| 58–61 | 6.8 ± 0.1 ab | |||
| 61–64 | 3.5 ± 1.7 a | |||
| 64–67 | 3.9 ± 1.0 a | |||
| 67–70 | 3.3 ± 1.9 a | |||
| 70–73 | −4.2 ± 2.5 c | |||
| Enzyme | Specific Enzyme Activity (U/mg protein) | ||
|---|---|---|---|
| Free Cells | Immobilized Cells 15th Day | ||
| 15th Day | 35th Day | ||
| O-demethylase | 412.84 ± 48.53 a | 737.16 ± 55.81 b | 1051.84 ± 65.57 c |
| Aromatic monooxygenase (Phe) | 13.06 ± 0.83 a | 27.14 ± 2.40 b | 31.55 ± 1.18 c |
| Aromatic monooxygenase (Npx) | 14.78 ± 1.28 a | 65.17 ± 3.59 b | 123.71 ± 12.39 c |
| Naphthalene dioxygenase | 8.16 ± 0.82 a | 10.71 ± 2.23 a | 15.73 ± 1.80 b |
| Gentisate 1,2-dioxygenase | 52.95 ± 2.90 a | 122.26 ± 9.44 b | 203.03 ± 18.55 c |
| Salicylate 1,2-dioxygenase | 388.26 ± 11.12 a | 520.38 ± 24.60 b | 714.87 ± 71.58 c |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dzionek, A.; Wojcieszyńska, D.; Hupert-Kocurek, K.; Adamczyk-Habrajska, M.; Guzik, U. Immobilization of Planococcus sp. S5 Strain on the Loofah Sponge and Its Application in Naproxen Removal. Catalysts 2018, 8, 176. https://doi.org/10.3390/catal8050176
Dzionek A, Wojcieszyńska D, Hupert-Kocurek K, Adamczyk-Habrajska M, Guzik U. Immobilization of Planococcus sp. S5 Strain on the Loofah Sponge and Its Application in Naproxen Removal. Catalysts. 2018; 8(5):176. https://doi.org/10.3390/catal8050176
Chicago/Turabian StyleDzionek, Anna, Danuta Wojcieszyńska, Katarzyna Hupert-Kocurek, Małgorzata Adamczyk-Habrajska, and Urszula Guzik. 2018. "Immobilization of Planococcus sp. S5 Strain on the Loofah Sponge and Its Application in Naproxen Removal" Catalysts 8, no. 5: 176. https://doi.org/10.3390/catal8050176
APA StyleDzionek, A., Wojcieszyńska, D., Hupert-Kocurek, K., Adamczyk-Habrajska, M., & Guzik, U. (2018). Immobilization of Planococcus sp. S5 Strain on the Loofah Sponge and Its Application in Naproxen Removal. Catalysts, 8(5), 176. https://doi.org/10.3390/catal8050176