Three Multi-Components Reaction: Synthesis and X-Ray Single-Crystal of Hydroacridinone-Based Hydrazino-S-Triazine Derivative as a New Class of Urease Inhibitor
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Methods
2.2. Synthesis of Hydroacridinone Compound 4
2.3. Single-Crystal X-ray Diffraction Analysis
2.4. Hirshfeld Surface Analysis
2.5. Computational Methods
2.6. Urease Inhibition
3. Results and Discussion
3.1. Chemistry
3.2. Crystal Structural Description
3.3. Crystal Packing
3.4. Hirshfeld Analysis of Molecular Packing
3.5. Geometric Parameters
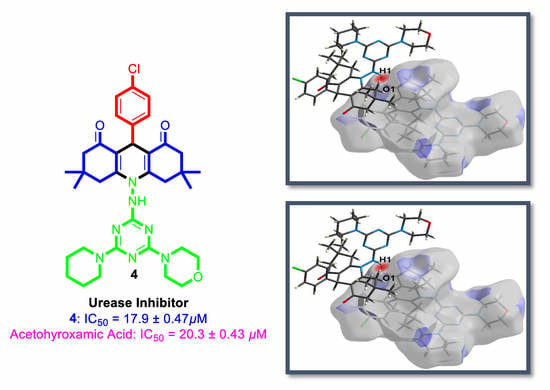
3.6. Biological Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hameed, A.; Al-Rashida, M.; Uroos, M.; Qazi, S.U.; Naz, S.; Ishtiaq, M.; Khan, K.M. A patent update on therapeutic applications of urease inhibitors (2012–2018). Expert Opin. Ther. Pat. 2019, 29, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Rego, Y.F.; Queiroz, M.P.; Brito, T.O.; Carvalho, P.G.; de Queiroz, V.T.; de Fatima, A.; Macedo, F., Jr. A review on the development of urease inhibitors as antimicrobial agents against pathogenic bacteria. J. Adv. Res. 2018, 13, 69–100. [Google Scholar] [CrossRef] [PubMed]
- Sebestik, J.; Hlavacek, J.; Stibor, I. A role of the 9-aminoacridines and their conjugates in a life science. Curr. Protein Pept. Sci. 2007, 8, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Huo, L.; Jaiswal, Y.; Huang, J.; Zhong, Z.; Zhong, J.; Williams, L.; Xia, X.; Liang, Y.; Yan, Z. Design, Synthesis, Antimicrobial, and Anticancer Activities of Acridine Thiosemicarbazides Derivatives. Molecules 2019, 24, 2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, M.O.; Sherrill, J.; Madrid, P.B.; Liou, A.P.; Weisman, J.L.; DeRisi, J.L.; Guy, R.K. Parallel synthesis of 9-aminoacridines and their evaluation against chloroquine-resistant Plasmodium falciparum. Bioorganic Med. Chem. 2006, 14, 334–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luedtke, N.W.; Liu, Q.; Tor, Y. RNA—Ligand interactions: Affinity and specificity of aminoglycoside dimers and acridine conjugates to the HIV-1 Rev response element. Biochemistry 2003, 42, 11391–11403. [Google Scholar] [CrossRef]
- Sondhi, S.M.; Singh, J.; Rani, R.; Gupta, P.P.; Agrawal, S.K.; Saxena, A.K. Synthesis, anti-inflammatory and anticancer activity evaluation of some novel acridine derivatives. Eur. J. Med. Chem. 2010, 45, 555–563. [Google Scholar] [CrossRef]
- Bondinell, W.; Reader, V.; Ku, T. SmithKline Beecham Corp, Substituted Bis-Acridines and Related Compounds as CCR5 Receptor Ligands, Anti-Inflammatory Agents and Anti-Viral Agents. U.S. Patent Application 09/833,044, 18 October 2001. [Google Scholar]
- Belmont, P.; Bosson, J.; Godet, T.; Tiano, M. Acridine and acridone derivatives, anticancer properties and synthetic methods: Where are we now? Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem.-Anti-Cancer Agents) 2007, 7, 139–169. [Google Scholar] [CrossRef]
- Denny, W.A.; Pereira, R.C.; Pontinha, A.D.R.; Pineiro, M.; de Melo, J.S.S. A comprehensive spectral, photophysical and electrochemical study of synthetic water-soluble acridones. A new class of pH and polarity sensitive fluorescent probes. Dye. Pigment. 2019, 166, 203–210. [Google Scholar]
- Denny, W.A. Acridine derivatives as chemotherapeutic agents. Curr. Med. Chem. 2002, 9, 1655–1665. [Google Scholar] [CrossRef]
- Zhang, B.; Li, X.; Li, B.; Gao, C.; Jiang, Y. Acridine and its derivatives: A patent review (2009–2013). Expert Opin. Ther. Pat. 2014, 24, 647–664. [Google Scholar] [CrossRef] [PubMed]
- Belmont, P.; Dorange, I. Acridine/acridone: A simple scaffold with a wide range of application in oncology. Expert Opin. Ther. Pat. 2008, 18, 1211–1224. [Google Scholar] [CrossRef]
- Galdino-Pitta, M.R.; Pitta, M.G.R.; Lima, M.C.A.; Galdino, L.S.; Pitta, R.I. Niche for acridine derivatives in anticancer therapy. Mini Rev. Med. Chem. 2013, 13, 1256–1271. [Google Scholar] [PubMed]
- Kaur, J.; Singh, P. Acridine derivatives: A patent review (2009–2010). Expert Opin. Ther. Pat. 2011, 21, 437–454. [Google Scholar] [CrossRef] [PubMed]
- Ballell, L.; Field, R.A.; Duncan, K.; Young, R.J. New small-molecule synthetic antimycobacterials. Antimicrob. Agents Chemother. 2005, 49, 2153–2163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, D.P.; Chang, Y.T. Chemical genetics. Chem. Rev. 2006, 106, 2476–2530. [Google Scholar] [CrossRef]
- Khattab, S.; Khalil, H.; Bekhit, A.; El-Rahman, M.; El-Faham, A.; Albericio, F. Synthesis and preliminary biological evaluation of 1,3,5-triazine amino acid derivatives to study their MAO inhibitors. Molecules 2015, 20, 15976–15988. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.Z.; Chen, F.E.; Balzarini, J.; De Clercq, E.; Pannecouque, C. Non-nucleoside HIV-1 reverse transcriptase inhibitors. Part 11: Structural modulations of diaryltriazines with potent anti-HIV activity. Eur. J. Med. Chem. 2008, 43, 1230–1236. [Google Scholar] [CrossRef]
- Saleh, M.; Abbott, S.; Perron, V.; Lauzon, C.; Penney, C.; Zacharie, B. Synthesis and antimicrobial activity of 2-fluorophenyl-4, 6-disubstituted [1,3,5] triazines. Bioorganic Med. Chem. Lett. 2010, 20, 945–949. [Google Scholar] [CrossRef]
- Khattab, S.N.; Naim, S.E.A.; El-Sayed, M.; El Bardan, A.A.; Elzoghby, A.O.; Bekhit, A.A.; El-Faham, A. Design and synthesis of new s-triazine polymers and their application as nanoparticulate drug delivery systems. New J. Chem. 2016, 40, 9565–9578. [Google Scholar] [CrossRef]
- Zhou, C.; Min, J.; Liu, Z.; Young, A.; Deshazer, H.; Gao, T.; Chang, Y.T.; Kallenbach, N.R. Synthesis and biological evaluation of novel 1,3,5-triazine derivatives as antimicrobial agents. Bioorganic Med. Chem. Lett. 2008, 18, 1308–1311. [Google Scholar] [CrossRef] [PubMed]
- Melato, S.; Prosperi, D.; Coghi, P.; Basilico, N.; Monti, D. A combinatorial approach to 2, 4, 6-trisubstituted triazines with potent antimalarial activity: Combining conventional synthesis and microwave-assistance. ChemMedChem Chem. Enabling Drug Discov. 2008, 3, 873–876. [Google Scholar]
- Nowakowska, Z. A review of anti-infective and anti-inflammatory chalcones. Eur. J. Med. Chem. 2007, 42, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Haiba, N.S.; Khalil, H.H.; Moniem, M.A.; El-Wakil, M.H.; Bekhit, A.A.; Khattab, S.N. Design, synthesis and molecular modeling studies of new series of s-triazine derivatives as antimicrobial agents against multi-drug resistant clinical isolates. Bioorganic Chem. 2019, 89, 103013. [Google Scholar] [CrossRef]
- Khattab, S.N.; Khalil, H.H.; Bekhit, A.A.; Abd El-Rahman, M.M.; de la Torre, B.G.; El-Faham, A.; Albericio, F. 1,3,5-Triazino peptide derivatives: Synthesis, characterization, and preliminary antileishmanial activity. ChemMedChem 2018, 13, 725–735. [Google Scholar] [CrossRef] [Green Version]
- Gavade, S.N.; Markad, V.L.; Kodam, K.M.; Shingare, M.S.; Mane, D.V. Synthesis and biological evaluation of novel 2, 4, 6-triazine derivatives as antimicrobial agents. Bioorganic Med. Chem. Lett. 2012, 22, 5075–5077. [Google Scholar] [CrossRef]
- Courme, C.; Gresh, N.; Vidal, M.; Lenoir, C.; Garbay, C.; Florent, J.C.; Bertounesque, E. Synthesis of aryl phosphates based on pyrimidine and triazine scaffolds. Eur. J. Med. Chem. 2010, 45, 244–255. [Google Scholar] [CrossRef]
- Gahtori, P.; Ghosh, S.K.; Singh, B.; Singh, U.P.; Bhat, H.R.; Uppal, A. Synthesis, SAR and antibacterial activity of hybrid chloro, dichloro-phenylthiazolyl-s-triazines. Saudi Pharm. J. 2012, 20, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Desai, N.C.; Makwana, A.H.; Rajpara, K.M. Synthesis and study of 1, 3, 5-triazine based thiazole derivatives as antimicrobial agents. J. Saudi Chem. Soc. 2016, 20, S334–S341. [Google Scholar] [CrossRef] [Green Version]
- Bruker, A. SAINT Software Reference Manual; Technical Publications Department: Madison, WI, USA, 1998; p. 5465. [Google Scholar]
- Spek, A.L.J. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer17; University of Western Australia: Perth, Australia, 2017; Available online: http://hirshfeldsurface.net (accessed on 30 December 2019).
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. GAUSSIAN 09. Revision A0; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Keith, T.; Millam, J. GaussView, Version 4.1; Dennington, R., Ed.; Semichem Inc.: Shawnee Mission, KS, USA, 2007. [Google Scholar]
- Weatherburn, M.W. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 1967, 39, 971–974. [Google Scholar] [CrossRef]
- Benini, S.; Rypniewski, W.R.; Wilson, K.S.; Miletti, S.; Ciurli, S.; Mangani, S. The complex of Bacillus pasteurii urease with acetohydroxamate anion from X-ray data at 1.55 Å resolution. JBIC J. Biol. Inorg. Chem. 2000, 5, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Maheswari, C.S.; Sivaguru, P.; Grzegorz Malecki, J.; Lalitha, A. Glacial acetic acid-assisted one-pot synthesis of diverse octahydroacridin-4-methylbenzenesulfonamides via Tandem Cascade reactions. Polycycl. Aromat. Compd. 2018, 1–14. [Google Scholar] [CrossRef]
- Cremer, D.T.; Pople, J.A. General definition of ring puckering coordinates. J. Am. Chem. Soc. 1975, 97, 1354–1358. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 3814–3816. [Google Scholar] [CrossRef]
- Geerlings, P.; De Proft, F.; Langenaeker, W. Conceptual Density Functional Theory. Chem. Rev. 2003, 103, 1793. [Google Scholar] [CrossRef]
- Domingo, L.R.; Ríos-Gutiérrez, M.; Pérez, P. Applications of the conceptual density functional theory indices to organic chemistry reactivity. Molecules 2016, 21, 748. [Google Scholar] [CrossRef] [Green Version]
- Foresman, J.B.; Frisch, A. Exploring Chemistry with Electronic Structure Methods, 2nd ed.; Gaussian: Pittsburgh, PA, USA, 1996. [Google Scholar]
- Chang, R. Chemistry, 7th ed.; McGraw-Hill: New York, NY, USA, 2001. [Google Scholar]
- Kosar, B.; Albayrak, C. Spectroscopic investigations and quantum chemical computational study of (E)-4-methoxy-2-[(p-tolylimino)methyl]phenol. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 78, 160–167. [Google Scholar] [CrossRef]
- Koopmans, T.A. Über die Zuordnung von Wellenfunktionen und Eigenwerten zu den einzelnen Elektronen eines Atoms. Physica 1934, 1, 104–113. [Google Scholar] [CrossRef]
- Parr, R.G.; Yang, W. Density Functional Theory of Atoms and Molecules; Oxford University Press: Oxford, UK, 1989. [Google Scholar]
- Parr, R.G.; Szentpaly, L.V.; Liu, S. Electrophilicity index. J. Am. Chem. Soc. 1999, 121, 1922–1924. [Google Scholar] [CrossRef]
- Domingo, L.R.; Chamorro, E.; Pérez, P. Understanding the reactivity of captodative ethylenes in polar cycloaddition reactions. A Theoretical study. J. Org. Chem. 2008, 73, 4615. [Google Scholar] [CrossRef] [PubMed]
- El-Faham, A.; Soliman, S.M.; Ghabbour, H.A.; Elnakady, Y.A.; Mohaya, T.A.; Siddiqui, M.R.; Albericio, F. Ultrasonic promoted synthesis of novel s-triazine-Schiff base derivatives; molecular structure, spectroscopic studies and their preliminary anti-proliferative activities. J. Mol. Struct. 2016, 1125, 121–135. [Google Scholar] [CrossRef]
- Farooq, M.; Sharma, A.; Almarhoon, Z.; Al-Dhfyan, A.; El-Faham, A.; Taha, N.A.; Wadaan, M.A.; Beatriz, G.; Albericio, F. Design and synthesis of mono-and di-pyrazolyl-s-triazine derivatives, their anticancer profile in human cancer cell lines, and in vivo toxicity in zebrafish embryos. Bioorganic Chem. 2019, 87, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.R.; Modh, R.P.; Chikhalia, K.H. Privileged s-triazines: Structure and pharmacological applications. Future Med. Chem. 2014, 6, 463–477. [Google Scholar] [CrossRef]










| Identification Code | 4 |
|---|---|
| Empirical formula | C35 H44 Cl N7 O3 |
| Formula weight | 646.22 |
| Temperature | 173(2) K |
| Wavelength | 1.54178 Å |
| Crystal system | Orthorhombic |
| Space group | Pbca |
| Unit cell dimensions | a = 11.6271(2) Å α = 90°. b = 18.2018(4) Å β = 90°. c = 32.4721(6) Å γ = 90°. |
| Volume | 6872.2(2) Å3 |
| Z | 8 |
| Density (calculated) | 1.249 Mg/m3 |
| Absorption coefficient | 1.343 mm−1 |
| F(000) | 2752 |
| Crystal size | 0.150 × 0.090 × 0.070 mm3 |
| Theta range for data collection | 2.721° to 68.235°. |
| Index ranges | −14 ≤ h ≤ 14, −20 ≤ k ≤ 21, −39 ≤ l ≤ 38 |
| Reflections collected | 51121 |
| Independent reflections | 6266 [R(int) = 0.0788] |
| Completeness to theta = 67.679° | 99.9% |
| Refinement method | Full-matrix least-squares on F2 |
| Data / restraints / parameters | 6266 / 0 / 423 |
| Goodness-of-fit on F2 | 1.026 |
| Final R indices [I>2sigma(I)] | R1 = 0.0561, wR2 = 0.1457 |
| R indices (all data) | R1 = 0.0814, wR2 = 0.1630 |
| Largest diff. peak and hole | 0.278 and −0.418 e.Å−3 |
| CCDC No.1967280 | |
| D-H…A | D-H | H…A | D…A | D-H…A |
|---|---|---|---|---|
| N2 -H1...O1(i) | 0.89(2) | 1.93(3) | 2.727(3) | 148(2) |
| C12-H12B…C31(ii) | 0.99 | 2.898 | 3.813(3) | 153.9 |
| N2-H1…C4(iii) | 0.89(2) | 2.73(3) | 3.573(3) | 159(2) |
| Compound | Urease Inhibition IC50 ± SEM (µM) |
|---|---|
| 4 | 17.9 ± 0.47 |
| Acetohyroxamic Acid | 20.3 ± 0.43 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barakat, A.; Soliman, S.M.; El-Faham, A.; Ali, M.; Al-Majid, A.M.; Yousuf, S.; Choudhary, M.I. Three Multi-Components Reaction: Synthesis and X-Ray Single-Crystal of Hydroacridinone-Based Hydrazino-S-Triazine Derivative as a New Class of Urease Inhibitor. Crystals 2020, 10, 14. https://doi.org/10.3390/cryst10010014
Barakat A, Soliman SM, El-Faham A, Ali M, Al-Majid AM, Yousuf S, Choudhary MI. Three Multi-Components Reaction: Synthesis and X-Ray Single-Crystal of Hydroacridinone-Based Hydrazino-S-Triazine Derivative as a New Class of Urease Inhibitor. Crystals. 2020; 10(1):14. https://doi.org/10.3390/cryst10010014
Chicago/Turabian StyleBarakat, Assem, Saied M. Soliman, Ayman El-Faham, M. Ali, Abdullah Mohammed Al-Majid, Sammer Yousuf, and M. Iqbal Choudhary. 2020. "Three Multi-Components Reaction: Synthesis and X-Ray Single-Crystal of Hydroacridinone-Based Hydrazino-S-Triazine Derivative as a New Class of Urease Inhibitor" Crystals 10, no. 1: 14. https://doi.org/10.3390/cryst10010014
APA StyleBarakat, A., Soliman, S. M., El-Faham, A., Ali, M., Al-Majid, A. M., Yousuf, S., & Choudhary, M. I. (2020). Three Multi-Components Reaction: Synthesis and X-Ray Single-Crystal of Hydroacridinone-Based Hydrazino-S-Triazine Derivative as a New Class of Urease Inhibitor. Crystals, 10(1), 14. https://doi.org/10.3390/cryst10010014