Inhibition of Tumor Growth via Systemic siRNA Delivery Using Reducible Bile Acid-Conjugated Polyethylenimine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Small Interfering RNAs (siRNAs) Design
2.3. Synthesis of DA-PEI and TP-DA-PEI Conjugates
2.4. Preparation of TP-DA-PEI/thiol-siRNA
2.5. Serum Stability
2.6. In Vitro Gene Silencing Test of TP-DA-PEI/thiol-siRNA
2.7. Cytotoxicity Assay
2.8. Intracellular Uptake of TP-DA-PEI/thiol-siRNA
2.9. In Vivo Imaging
2.10. Tumor Growth Inhibition
3. Results and Discussion
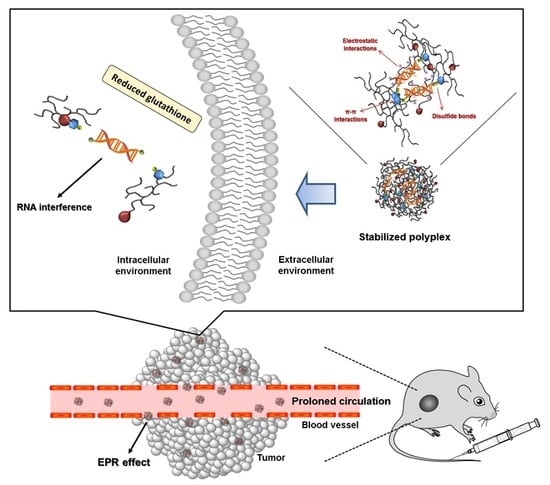
3.1. Formation and Characterization of TP-DA-PEI/thiol-siRNA
3.2. In Vitro Gene Silencing Efficiency of TP-DA-PEI/thiol-siRNA
3.3. Serum Stability and In Vitro Gene Silencing of TP-DA-PEI/thiol-siRNA
3.4. Intracellular Uptake of TP-DA-PEI/thiol-siRNA
3.5. In Vivo Biodistribution and Anti-Tumor Effect of TP-DA-PEI/thiol-siRNA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Devi, G.R. Sirna-based approaches in cancer therapy. Cancer Gene Ther. 2006, 13, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E.; Zuckerman, J.E.; Choi, C.H.J.; Seligson, D.; Tolcher, A.; Alabi, C.A.; Yen, Y.; Heidel, J.D.; Ribas, A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizrahy, S.; Hazan-Halevy, I.; Dammes, N.; Landesman-Milo, D.; Peer, D. Current progress in non-viral RNAi-based delivery strategies to lymphocytes. Mol. Ther. 2017, 25, 1491–1500. [Google Scholar] [CrossRef] [PubMed]
- Peer, D.; Lieberman, J. Special delivery: Targeted therapy with small RNAs. Gene Ther. 2011, 18, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.H.; Kim, S.W.; Park, T.G. Molecular design of functional polymers for gene therapy. Prog. Polym. Sci. 2007, 32, 1239–1274. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.; Bauman, J.; Kang, H.; Ming, X. Biological barriers to therapy with antisense and siRNA oligonucleotides. Mol. Pharm. 2009, 6, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Muthiah, M.; Chellasamy, G.; Natarajan, R.; Subramanian, S.; Chinnappa, S. Proton conducting polymer electrolytes based on PVdF-PVA with NH4NO3. J. Polym. Eng. 2013, 33, 315–322. [Google Scholar] [CrossRef]
- Ninan, N.; Muthiah, M.; Park, I.K.; Elain, A.; Wong, T.W.; Thomas, S.; Grohens, Y. Faujasites incorporated tissue engineering scaffolds for wound healing: In vitro and in vivo analysis. ACS Appl. Mater. Interfaces 2013, 5, 11194–11206. [Google Scholar] [CrossRef] [PubMed]
- Worgall, S.; Wolff, G.; FalckPedersen, E.; Crystal, R.G. Innate immune mechanisms dominate elimination of adenoviral vectors following in vivo administration. Hum. Gene Ther. 1997, 8, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Mok, H.; Lee, S.H.; Park, J.W.; Park, T.G. Multimeric small interfering ribonucleic acid for highly efficient sequence-specific gene silencing. Nat. Mater. 2010, 9, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.C.; Bae, Y.H. Co-delivery of small interfering RNA and plasmid DNA using a polymeric vector incorporating endosomolytic oligomeric sulfonamide. Biomaterials 2011, 32, 4914–4924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, H.H.; Joo, M.K.; Mok, H.; Lee, M.; Hwang, K.C.; Kim, S.W.; Jeong, J.H.; Choi, D.; Kim, S.H. MSC-based VEGF gene therapy in rat myocardial infarction model using facial amphipathic bile acid-conjugated polyethyleneimine. Biomaterials 2014, 35, 1744–1754. [Google Scholar] [CrossRef] [PubMed]
- Chae, S.Y.; Kim, H.J.; Lee, M.S.; Jang, Y.L.; Lee, Y.; Lee, S.H.; Lee, K.; Kim, S.H.; Kim, H.T.; Chi, S.C.; et al. Energy-independent intracellular gene delivery mediated by polymeric biomimetics of cell-penetrating peptides. Macromol. Biosci. 2011, 11, 1169–1174. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, D.; Jang, Y.L.; Chae, S.Y.; Choi, D.; Jeong, J.H.; Kim, S.H. Facial amphipathic deoxycholic acid-modified polyethyleneimine for efficient mmp-2 siRNA delivery in vascular smooth muscle cells. Eur. J. Pharm. Biopharm. 2012, 81, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Hong, J.; Moon, H.H.; Nam, H.Y.; Mok, H.; Jeong, J.H.; Kim, S.W.; Choi, D.; Kim, S.H. Anti-apoptotic cardioprotective effects of shp-1 gene silencing against ischemia-reperfusion injury: Use of deoxycholic acid-modified low molecular weight polyethyleneimine as a cardiac siRNA-carrier. J. Control. Release 2013, 168, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Ku, S.H.; Lee, M.S.; Jeong, J.H.; Mok, H.; Choi, D.; Kim, S.H. Cardiac RNAi therapy using rage sirna/deoxycholic acid-modified polyethylenimine complexes for myocardial infarction. Biomaterials 2014, 35, 7562–7573. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Ku, S.H.; Park, H.; Hong, J.; Kim, D.; Choi, B.R.; Pak, H.N.; Lee, M.H.; Mok, H.; Jeong, J.H.; et al. Rage siRNA-mediated gene silencing provides cardioprotection against ventricular arrhythmias in acute ischemia and reperfusion. J. Control. Release 2015, 217, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Son, S.; Son, S.; Kim, N.; Yhee, J.Y.; Lee, J.H.; Choi, J.S.; Joo, C.K.; Lee, H.; Lee, D.; et al. Reducible polyethylenimine nanoparticles for efficient siRNA delivery in corneal neovascularization therapy. Macromol. Biosci. 2016, 16, 1583–1597. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.L.; Ku, S.H.; Lee, S.J.; Park, J.H.; Kim, W.J.; Kwon, I.C.; Kim, S.H.; Jeong, J.H. Hyaluronic acid-siRNA conjugate/reducible polyethylenimine complexes for targeted siRNA delivery. J. Nanosci. Nanotechnol. 2014, 14, 7388–7394. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.L.; Yun, U.J.; Lee, M.S.; Kim, M.G.; Son, S.; Lee, K.; Chae, S.Y.; Lim, D.W.; Kim, H.T.; Kim, S.H.; et al. Cell-penetrating peptide mimicking polymer-based combined delivery of paclitaxel and siRNA for enhanced tumor growth suppression. Int. J. Pharm. 2012, 434, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; van Steenbergen, M.J.; Teunissen, E.A.; Novo, L.; Gradmann, S.; Baldus, M.; van Nostrum, C.F.; Hennink, W.E. Pi-pi stacking increases the stability and loading capacity of thermosensitive polymeric micelles for chemotherapeutic drugs. Biomacromolecules 2013, 14, 1826–1837. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Lee, K.D. Enhanced gene delivery using disulfide-crosslinked low molecular weight polyethylenimine with listeriolysin o-polyethylenimine disulfide conjugate. J. Control. Release 2008, 131, 70–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinosa-Diez, C.; Miguel, V.; Mennerich, D.; Kietzmann, T.; Sanchez-Perez, P.; Cadenas, S.; Lamas, S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015, 6, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.C.; Khosla, C. Thiol-disulfide exchange reactions in the mammalian extracellular environment. Annu. Rev. Chem. Biomol. 2016, 7, 197–222. [Google Scholar] [CrossRef] [PubMed]
- Rhee, J.; Hoff, P.M. Angiogenesis inhibitors in the treatment of cancer. Expert Opin. Pharmacother. 2005, 6, 1701–1711. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jeong, J.H.; Lee, S.H.; Kim, S.W.; Park, T.G. Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J. Control. Release 2008, 129, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, Z.; Li, Y. Dual-degradable disulfide-containing pei-pluronic/DNA polyplexes: Transfection efficiency and balancing protection and DNA release. Int. J. Nanomed. 2013, 8, 3689–3701. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Sun, Y.; Xian, L.; Xun, Z.; Yu, J.; Yang, T.; Zhao, X.; Cai, C.; Wang, D.; Ding, P. Disulfide-bond-containing agamatine-cystaminebisacrylamide polymer demonstrates better transfection efficiency and lower cytotoxicity than polyethylenimine in NIH/3T3 cells. J. Cell. Biochem. 2018, 119, 1767–1779. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Wu, J.; Sawa, T.; Matsumura, Y.; Hori, K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: A review. J. Control. Release 2000, 65, 271–284. [Google Scholar] [CrossRef]









| Polymer | Molar Feed Ratio (DA-PEI/FTP) | Composition 1 (DA-PEI/FTP) |
|---|---|---|
| TP(1)-DA-PEI | 1:1 | 1:0.81 |
| TP(3)-DA-PEI | 1:3 | 1:2.13 |
| TP(5)-DA-PEI | 1:5 | 1:3.14 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, Y.; Lee, J.E.; Kim, N.W.; Lee, J.H.; Lim, S.Y.; Kim, E.S.; Park, J.W.; Lee, M.S.; Jeong, J.H. Inhibition of Tumor Growth via Systemic siRNA Delivery Using Reducible Bile Acid-Conjugated Polyethylenimine. Polymers 2018, 10, 953. https://doi.org/10.3390/polym10090953
Yin Y, Lee JE, Kim NW, Lee JH, Lim SY, Kim ES, Park JW, Lee MS, Jeong JH. Inhibition of Tumor Growth via Systemic siRNA Delivery Using Reducible Bile Acid-Conjugated Polyethylenimine. Polymers. 2018; 10(9):953. https://doi.org/10.3390/polym10090953
Chicago/Turabian StyleYin, Yue, Jung Eun Lee, Nak Won Kim, Jong Han Lee, Su Yeon Lim, E Seul Kim, Ji Won Park, Min Sang Lee, and Ji Hoon Jeong. 2018. "Inhibition of Tumor Growth via Systemic siRNA Delivery Using Reducible Bile Acid-Conjugated Polyethylenimine" Polymers 10, no. 9: 953. https://doi.org/10.3390/polym10090953
APA StyleYin, Y., Lee, J. E., Kim, N. W., Lee, J. H., Lim, S. Y., Kim, E. S., Park, J. W., Lee, M. S., & Jeong, J. H. (2018). Inhibition of Tumor Growth via Systemic siRNA Delivery Using Reducible Bile Acid-Conjugated Polyethylenimine. Polymers, 10(9), 953. https://doi.org/10.3390/polym10090953