Structural Polymorphism of Single pDNA Condensates Elicited by Cationic Block Polyelectrolytes
Abstract
:1. Introduction
2. DNA Condensation Induced by Low- or High-Molecular-Weight Polycations
3. Single-DNA Condensation Regulated by Copolymers
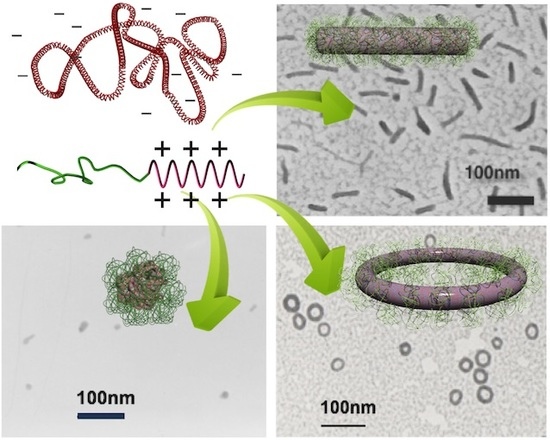
3.1. Globular, Rod-Shaped, and Toroidal Structures
3.2. Arrangements of DNA Strands in Rod-Shaped and Toroidal PMs
3.3. Folding Mechanism of DNA in PMs and Their Structural Polymorphism
4. PMs as Potential Gene Vectors
5. Summary and Outlook
Funding
Conflicts of Interest
References
- Minagawa, K.; Matsuzawa, Y.; Yoshikawa, K.; Matsumoto, M.; Doi, M. Direct observation of the biphasic conformational change of DNA induced by cationic polymers. FEBS Lett. 1991, 295, 67–69. [Google Scholar] [CrossRef] [Green Version]
- Bloomfield, V.A. DNA condensation by multivalent cations. Biopolymers 1997, 44, 269–282. [Google Scholar] [CrossRef]
- Sergeyev, V.G.; Pyshkina, O.A.; Lezov, A.V.; Mel’nikov, A.B.; Ryumtsev, E.I.; Zezin, A.B.; Kabanov, V.A. DNA complexed with oppositely charged amphiphile in low-polar organic solvents. Langmuir 1999, 15, 4434–4440. [Google Scholar] [CrossRef]
- Mel’nikov, S.M.; Sergeyev, V.G.; Yoshikawa, K. Discrete coil-globule transition of large DNA induced by cationic surfactant. J. Am. Chem. Soc. 1995, 117, 2401–2408. [Google Scholar] [CrossRef]
- Yoshikawa, K. Controlling the higher-order structure of giant DNA molecules. Adv. Drug Deliv. Rev. 2001, 52, 235–244. [Google Scholar] [CrossRef]
- Teif, V.B.; Bohinc, K. Condensed DNA: Condensing the concepts. Prog. Biophys. Mol. Biol. 2011, 105, 208–222. [Google Scholar] [CrossRef]
- Smith, S.B.; Cui, Y.; Bustamante, C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 1996, 271, 795–799. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Weers, B.; Stellwagen, N.C. DNA persistence length revisited. Biopolymers 2001, 61, 261–275. [Google Scholar] [CrossRef]
- Sim, A.Y.L. Nucleic acid polymeric properties and electrostatics: Directly comparing theory and simulation with experiment. Adv. Colloid Interface Sci. 2016, 232, 49–56. [Google Scholar] [CrossRef]
- Shakya, A.; King, J.T. DNA Local-Flexibility-Dependent Assembly of Phase-Separated Liquid Droplets. Biophys. J. 2018, 115, 1840–1847. [Google Scholar] [CrossRef] [Green Version]
- Vieregg, J.R.; Lueckheide, M.; Marciel, A.B.; Leon, L.; Bologna, A.J.; Rivera, J.R.; Tirrell, M.V. Oligonucleotide-Peptide complexes: Phase control by hybridization. J. Am. Chem. Soc. 2018, 140, 1632–1638. [Google Scholar] [CrossRef]
- André, A.A.M.; Spruijt, E. Rigidity rules in DNA droplets: Nucleic acid flexibility affects model membraneless organelles. Biophys. J. 2018, 115, 1837–1839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kataoka, K.; Togawa, H.; Harada, A.; Yasugi, K.; Matsumoto, T.; Katayose, S. Spontaneous formation of polyion complex micelles with narrow distribution from antisense oligonucleotide and cationic block copolymer in physiological saline. Macromolecules 1996, 29, 8556–8557. [Google Scholar] [CrossRef]
- Wolfert, M.A.; Schacht, E.H.; Toncheva, V.; Ulbrich, K.; Nazarova, O.; Seymour, L.W. Characterization of vectors for gene therapy formed by self-assembly of DNA with synthetic block co-polymers. Hum. Gene Ther. 1996, 7, 2123–2133. [Google Scholar] [CrossRef] [PubMed]
- Katayose, S.; Kataoka, K. Water-Soluble polyion complex associates of dna and poly (ethylene glycol)-poly (L-lysine) block copolymer. Bioconjugate Chem. 1997, 8, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Kabanov, A.V.; Kabanov, V.A. Interpolyelectrolyte and block ionomer complexes for gene delivery: Physico-Chemical aspects. Adv. Drug Deliv. Rev. 1998, 30, 49–60. [Google Scholar] [CrossRef]
- Dash, P.R.; Toncheva, V.; Schacht, E.; Seymour, L.W. Synthetic polymers for vectorial delivery of DNA: Characterisation of polymer-DNA complexes by photon correlation spectroscopy and stability to nuclease degradation and disruption by polyanions in vitro. J. Control. Release 1997, 48, 269–276. [Google Scholar] [CrossRef]
- Voets, I.K.; de Keizer, A.; Cohen Stuart, M.A. Complex coacervate core micelles. Adv. Colloid Interface Sci. 2009, 147–148, 300–318. [Google Scholar] [CrossRef]
- Osada, K. Versatile DNA folding structures organized by cationic block copolymers. Polym. J. 2019, 51, 381–387. [Google Scholar] [CrossRef]
- Kataoka, K.; Harada, A.; Yukio, N. Block copolymer micelles for drug delivery: Design, characterization and biological significance. Adv. Drug Deliv. Rev. 2001, 47, 113–131. [Google Scholar] [CrossRef]
- Park, T.G.; Jeong, J.H.; Kim, S.W. Current status of polymeric gene delivery systems. Adv. Drug Deliv. Rev. 2006, 58, 467–486. [Google Scholar] [CrossRef]
- Bloomfield, V.A. Condensation of DNA by multivalent cations: Considerations on mechanism. Biopolymers 1991, 31, 1471–1481. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa, Y.; Yonezawa, Y.; Yoshikawa, K. Formation of nucleation center in single double-stranded DNA chain. Biochem. Biophys. Res. Commun. 1996, 225, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Record, M.T.; Lohman, M.L.; De Haseth, P. Ion effects on ligand-nucleic acid interactions. J. Mol. Biol. 1976, 107, 145–158. [Google Scholar] [CrossRef]
- Kabanov, V.A.; Sergeyev, V.G.; Pyshkina, O.A.; Zinchenko, A.A.; Zezin, A.B.; Joosten, J.G.H.; Brackman, J.; Yoshikawa, K. Interpolyelectrolyte complexes formed by DNA and astramol poly(propylene imine) dendrimers. Macromolecules 2000, 33, 9587–9593. [Google Scholar] [CrossRef]
- Fu, J.; Schlenoff, J.B. Driving Forces for Oppositely Charged Polyion Association in Aqueous Solutions: Enthalpic, Entropic, but Not Electrostatic. J. Am. Chem. Soc. 2016, 138, 980–990. [Google Scholar] [CrossRef]
- Starodoubtsev, S.G.; Yoshikawa, K. Intrachain segregation in single giant DNA molecules induced by poly(2-vinylpyrrolidone). J. Phys. Chem. 1996, 100, 19702–19705. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Yoshikawa, Y.; Koyama, Y.; Kanbe, T. Highly effective compaction of long duplex DNA induced by polyethylene glycol with pendant amino groups. J. Am. Chem. Soc. 1997, 119, 6473–6477. [Google Scholar] [CrossRef]
- Takagi, S.; Tsumoto, K.; Yoshikawa, K. Intra-Molecular phase segregation in a single polyelectrolyte chain. J. Chem. Phys. 2001, 114, 6942–6949. [Google Scholar] [CrossRef]
- Chen, Q.; Osada, K.; Pennisi, M.; Uchida, S.; Tockary, T.A.; Dirisala, A.; Li, Y.; Takeda, K.M.; Oniyanagi, S.; Itaka, K.; et al. A tadpole-shaped gene carrier with distinct phase segregation in a ternary polymeric micelle. Soft Matter 2015, 11, 2718–2722. [Google Scholar] [CrossRef]
- Akitaya, T.; Seno, A.; Nakai, T.; Hazemoto, N.; Murata, S.; Yoshikawa, K. Weak interaction induces an ON/OFF switch, whereas strong interaction causes gradual change: Folding transition of a long duplex DNA chain by poly-L-lysine. Biomacromolecules 2007, 8, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Golan, R.; Pietrasanta, L.I.; Hsieh, W.; Hansma, H.G. DNA toroids: Stages in condensation. Biochemistry 1999, 38, 14069–14076. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Matsuzawa, Y. Nucleation and Growth in Single DNA Molecules. J. Am. Chem. Soc. 1996, 118, 929–930. [Google Scholar] [CrossRef]
- Hud, N.V.; Downing, K.H.; Balhorn, R. A constant radius of curvature model for the organization of DNA in toroidal condensates. Proc. Natl. Acad. Sci. USA 1995, 92, 3581–3585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilfan, I.D.; Conwell, C.C.; Sarkar, T.; Hud, N.V. Time study of DNA condensate morphology: Implications regarding the nucleation, growth, and equilibrium populations of toroids and rods. Biochemistry 2006, 45, 8174–8183. [Google Scholar] [CrossRef]
- Hud, N.V.; Vilfan, I.D. Toroidal DNA condensates: Unraveling the fine structure and the role of nucleation in determining size. Annu. Rev. Biophys. Biomol. Struct. 2005, 34, 295–318. [Google Scholar] [CrossRef] [PubMed]
- Carnerup, A.M.; Ainalem, M.L.; Alfredsson, V.; Nylander, T. Condensation of DNA using poly(amido amine) dendrimers: Effect of salt concentration on aggregate morphology. Soft Matter 2011, 7, 760–768. [Google Scholar] [CrossRef]
- Kabanov, A.V.; Kabanov, V.A. DNA complexes with polycations for the delivery of genetic material into cells. Bioconjugate Chem. 1995, 6, 7–20. [Google Scholar] [CrossRef]
- Lam, J.K.W.; Ma, Y.; Armes, S.P.; Lewis, A.L.; Baldwin, T.; Stolnik, S. Phosphorylcholine-polycation diblock copolymers as synthetic vectors for gene delivery. J. Control. Release 2004, 100, 293–312. [Google Scholar] [CrossRef]
- Takeda, K.M.; Osada, K.; Tockary, T.A.; Dirisala, A.; Chen, Q.; Kataoka, K. Poly(ethylene glycol) crowding as critical factor to determine pDNA packaging scheme into polyplex micelles for enhanced gene expression. Biomacromolecules 2016, 18, 36–43. [Google Scholar] [CrossRef]
- Rackstraw, B.J.; Martin, A.L.; Stolnik, S.; Roberts, C.J.; Garnett, M.C.; Davies, M.C.; Tendler, S.J.B. Microscopic investigations into PEG-cationic polymer-induced DNA condensation. Langmuir 2001, 17, 3185–3193. [Google Scholar] [CrossRef]
- Oupický, D.; Carlisle, R.C.; Seymour, L.W. Triggered intracellular activation of disulfide crosslinked polyelectrolyte gene delivery complexes with extended systemic circulation in vivo. Gene Ther. 2001, 8, 713–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanderkerken, S.; Toncheva, V.; Elomaa, M.; Mannisto, M.; Ruponen, M.; Schacht, E.; Urtti, A. Structure–Activity relationships of poly(L-lysines): Effects of pegylation and molecular shape on physicochemical and biological properties in gene delivery. J. Control. Release 2002, 83, 169–182. [Google Scholar]
- Ziady, A.G.; Gedeon, C.R.; Miller, T.; Quan, W.; Payne, J.M.; Hyatt, S.L.; Fink, T.L.; Muhammad, O.; Oette, S.; Kowalczyk, T.; et al. Transfection of airway epithelium by stable PEGylated poly-L-lysine DNA nanoparticles in vivo. Mol. Ther. 2003, 8, 936–947. [Google Scholar] [CrossRef]
- Osada, K.; Yamasaki, Y.; Katayose, S.; Kataoka, K. A synthetic block copolymer regulates S1 nuclease fragmentation of supercoiled plasmid DNA. Angew. Chem.–Int. Ed. 2005, 44, 3544–3548. [Google Scholar] [CrossRef] [PubMed]
- Boylan, N.J.; Suk, J.S.; Lai, S.K.; Jelinek, R.; Boyle, M.P.; Cooper, M.J.; Hanes, J. Highly compacted DNA nanoparticles with low MW PEG coatings: In vitro, ex vivo and in vivo evaluation. J. Control. Release 2012, 157, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osawa, S.; Osada, K.; Hiki, S.; Dirisala, A.; Ishii, T.; Kataoka, K. Polyplex micelles with double-protective compartments of hydrophilic shell and thermoswitchable palisade of poly(oxazoline)-based block copolymers for promoted gene transfection. Biomacromolecules 2016, 17, 354–361. [Google Scholar] [CrossRef]
- Li, J.; Chen, Q.; Zha, Z.; Li, H.; Toh, K.; Dirisala, A.; Matsumoto, Y.Y.; Osada, K.; Kataoka, K.; Ge, Z. Ternary polyplex micelles with PEG shells and intermediate barrier to complexed DNA cores for efficient systemic gene delivery. J. Control. Release 2015, 209, 77–87. [Google Scholar] [CrossRef]
- Qian, Y.; Zha, Y.; Feng, B.; Pang, Z.; Zhang, B.; Sun, X.; Ren, J.; Zhang, C.; Shao, X.; Zhang, Q.; et al. PEGylated poly(2-(dimethylamino) ethyl methacrylate)/DNA polyplex micelles decorated with phage-displayed TGN peptide for brain-targeted gene delivery. Biomaterials 2013, 34, 2117–2129. [Google Scholar] [CrossRef]
- Tockary, T.A.; Osada, K.; Chen, Q.; Machitani, K.; Dirisala, A.; Uchida, S.; Nomoto, T.; Toh, K.; Matsumoto, Y.; Itaka, K.; et al. Tethered PEG crowdedness determining shape and blood circulation profile of polyplex micelle gene carriers. Macromolecules 2013, 46, 6585–6592. [Google Scholar] [CrossRef]
- Ge, Z.; Chen, Q.; Osada, K.; Liu, X.; Tockary, T.A.; Uchida, S.; Dirisala, A.; Ishii, T.; Nomoto, T.; Toh, K.; et al. Targeted gene delivery by polyplex micelles with crowded PEG palisade and cRGD moiety for systemic treatment of pancreatic tumors. Biomaterials 2014, 35, 3416–3426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tockary, T.A.; Osada, K.; Motoda, Y.; Hiki, S.; Chen, Q.; Takeda, K.M.; Dirisala, A.; Osawa, S.; Kataoka, K. Rod-to-Globule transition of pDNA/PEG-poly(L-lysine) polyplex micelles induced by a collapsed balance between DNA rigidity and PEG crowdedness. Small 2016, 12, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Qu, W.; Pan, D.; Ren, Y.; Williford, J.M.; Cui, H.; Luijten, E.; Mao, H.Q. Plasmid-Templated shape control of condensed DNA-block copolymer nanoparticles. Adv. Mater. 2013, 25, 227–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Osada, K.; Chen, Q.; Tockary, T.A.; Dirisala, A.; Takeda, K.M.; Uchida, S.; Nagata, K.; Itaka, K.; Kataoka, K. Toroidal packaging of pDNA into block ionomer micelles exerting promoted in vivo gene expression. Biomacromolecules 2015, 16, 2664–2671. [Google Scholar] [CrossRef] [Green Version]
- Zinchenko, A.; Hiramatsu, H.; Yamaguchi, H.; Kubo, K.; Murata, S.; Kanbe, T.; Hazemoto, N.; Yoshikawa, K.; Akitaya, T. Amino Acid Sequence of Oligopeptide Causes Marked Difference in DNA Compaction and Transcription. Biophys. J. 2019, 116, 1836–1844. [Google Scholar] [CrossRef]
- Osada, K.; Oshima, H.; Kobayashi, D.; Doi, M.; Enoki, M.; Yamasaki, Y.; Kataoka, K. Quantized folding of plasmid DNA condensed with block catiomer into characteristic rod structures promoting transgene efficacy. J. Am. Chem. Soc. 2010, 132, 12343–12348. [Google Scholar] [CrossRef]
- Wenner, J.R.; Williams, M.C.; Rouzina, I.; Bloomfield, V.A. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 2002, 82, 3160–3169. [Google Scholar] [CrossRef] [Green Version]
- Osada, K.; Shiotani, T.; Tockary, T.A.; Kobayashi, D.; Oshima, H.; Ikeda, S.; Christie, R.J.; Itaka, K.; Kataoka, K. Enhanced gene expression promoted by the quantized folding of pDNA within polyplex micelles. Biomaterials 2012, 33, 325–332. [Google Scholar] [CrossRef]
- Ruff, Y.; Moyer, T.; Newcomb, C.J.; Demeler, B.; Stupp, S.I. Precision templating with DNA of a virus-like particle with peptide nanostructures. J. Am. Chem. Soc. 2013, 135, 6211–6219. [Google Scholar] [CrossRef]
- Lueckheide, M.; Vieregg, J.R.; Bologna, A.J.; Leon, L.; Tirrell, M.V. Structure-Property relationships of oligonucleotide polyelectrolyte complex micelles. Nano Lett. 2018. [Google Scholar] [CrossRef]
- Cerritelli, M.E.; Cheng, N.; Rosenberg, A.H.; McPherson, C.E.; Booy, F.P.; Steven, A.C. Encapsidated conformation of bacteriophage T7 DNA. Cell 1997, 91, 271–280. [Google Scholar] [CrossRef] [Green Version]
- Leforestier, A.; Livolant, F. Structure of toroidal DNA collapsed inside the phage capsid. Proc. Natl. Acad. Sci. USA 2009, 106, 9157–9162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rädler, J.O.; Koltover, I.; Salditt, T.; Safinya, C.R. Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science 1997, 275, 810–814. [Google Scholar] [CrossRef] [Green Version]
- DeRouchey, J.; Parsegian, V.A.; Rau, D.C. Cation charge dependence of the forces driving DNA assembly. Biophys. J. 2010, 99, 2608–2615. [Google Scholar] [CrossRef] [Green Version]
- Derouchey, J.; Netz, R.R.; Rädler, J.O. Structural investigations of DNA-polycation complexes. Eur. Phys. J. E 2005, 16, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Tinland, B.; Pluen, A.; Sturm, J.; Weill, G. Persistence length of single-stranded DNA. Macromolecules 1997, 30, 5763–5765. [Google Scholar] [CrossRef]
- Laemmli, U.K. Characterization of DNA condensates induced by poly(ethylene oxide) and polylysine. Proc. Natl. Acad. Sci. USA 1975, 72, 4288–4292. [Google Scholar] [CrossRef] [Green Version]
- Tockary, T.A.; Foo, W.; Dirisala, A.; Chen, Q.; Uchida, S.; Osawa, S.; Mochida, Y.; Liu, X.; Kinoh, H.; Cabral, H.; et al. Single-Stranded DNA-packaged polyplex micelle as adeno-associated-virus-inspired compact vector to systemically target stroma-rich pancreatic cancer. ACS Nano 2019, 13, 12732–12742. [Google Scholar] [CrossRef]
- Molas, M.; Bartrons, R.; Perales, J.C. Single-Stranded DNA condensed with poly-L-lysine results in nanometric particles that are significantly smaller, more stable in physiological ionic strength fluids and afford higher efficiency of gene delivery than their double-stranded counterparts. Biochim. Biophys. Acta–Gen. Subj. 2002, 1572, 37–44. [Google Scholar] [CrossRef]
- Petros, R.A.; Desimone, J.M. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discov. 2010, 9, 615–627. [Google Scholar] [CrossRef]
- Nomoto, T.; Matsumoto, Y.; Miyata, K.; Oba, M.; Fukushima, S.; Nishiyama, N.; Yamasoba, T.; Kataoka, K. In situ quantitative monitoring of polyplexes and polyplex micelles in the blood circulation using intravital real-time confocal laser scanning microscopy. J. Control. Release 2011, 151, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Konstan, M.W.; Davis, P.B.; Wagener, J.S.; Hilliard, K.A.; Stern, R.C.; Milgram, L.J.H.; Kowalczyk, T.H.; Hyatt, S.L.; Fink, T.L.; Gedeon, C.R.; et al. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum. Gene Ther. 2004, 15, 1255–1269. [Google Scholar] [CrossRef] [PubMed]
- Davis, P.B.; Cooper, M.J. Vectors for airway gene delivery. AAPS J. 2007, 9, E11–E17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dash, P.R.; Read, M.L.; Barrett, L.B.; Wolfert, M.A.; Seymour, L.W. Factors affecting blood clearance and in vivo distribution of polyelectrolyte complexes for gene delivery. Gene Ther. 1999, 6, 643–650. [Google Scholar] [CrossRef] [Green Version]
- Baliaka, A.; Zarogoulidis, P.; Domvri, K.; Hohenforst-Schmidt, W.; Sakkas, A.; Huang, H.; Le Pivert, P.; Koliakos, G.; Koliakou, E.; Kouzi-Koliakos, K.; et al. Intratumoral gene therapy versus intravenous gene therapy for distant metastasis control with 2-Diethylaminoethyl-Dextran Methyl Methacrylate Copolymer Non-Viral Vector-p53. Gene Ther. 2014, 21, 158–167. [Google Scholar] [CrossRef]
- Cui, L.; Osada, K.; Imaizumi, A.; Kataoka, K.; Nakano, K. Feasibility of a subcutaneously administered block/homo-mixed polyplex micelle as a carrier for DNA vaccination in a mouse tumor model. J. Control. Release 2015, 206, 220–231. [Google Scholar] [CrossRef]
- Meng, F.; Hennink, W.E.; Zhong, Z. Reduction-Sensitive polymers and bioconjugates for biomedical applications. Biomaterials 2009, 30, 2180–2198. [Google Scholar] [CrossRef]
- Dirisala, A.; Osada, K.; Chen, Q.; Tockary, T.A.; Machitani, K.; Osawa, S.; Liu, X.; Ishii, T.; Miyata, K.; Oba, M.; et al. Optimized rod length of polyplex micelles for maximizing transfection efficiency and their performance in systemic gene therapy against stroma-rich pancreatic tumors. Biomaterials 2014, 35, 5359–5368. [Google Scholar] [CrossRef] [Green Version]
- Yoshinaga, N.; Ishii, T.; Naito, M.; Endo, T.; Uchida, S.; Cabral, H.; Osada, K.; Kataoka, K. Polyplex micelles with phenylboronate/gluconamide cross-linking in the core exerting promoted gene transfection through spatiotemporal responsivity to intracellular pH and ATP concentration. J. Am. Chem. Soc. 2017, 139, jacs.7b08816. [Google Scholar] [CrossRef]
- Takeda, K.M.; Yamasaki, Y.; Dirisala, A.; Ikeda, S.; Tockary, T.A.; Toh, K.; Osada, K.; Kataoka, K. Effect of shear stress on structure and function of polyplex micelles from poly(ethylene glycol)-poly(L-lysine) block copolymers as systemic gene delivery carrier. Biomaterials 2017, 126, 31–38. [Google Scholar] [CrossRef]
- Ruponen, M.; Urtti, A. Interactions of polymeric and liposomal gene delivery system with extracellular glycosaminoglicans: Phisicochemical and transfection studies. Biochem. Biophys. Acta 1999, 1415, 331–341. [Google Scholar] [CrossRef] [Green Version]
- Burke, R.S.; Pun, S.H. Extracellular barriers to in vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjugate Chem. 2008, 19, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E. Effects of membrane-active agents in gene delivery. J. Control. Release 1998, 53, 155–158. [Google Scholar] [CrossRef]
- Wickham, T.J. Ligand-Directed targeting of genes to the site of disease. Nat. Med. 2003, 9, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Høgset, A.; Prasmickaite, L.; Selbo, P.K.; Hellum, M.; Engesaeter, B.; Bonsted, A.; Berg, K. Photochemical internalisation in drug and gene delivery. Adv. Drug Deliv. Rev. 2004, 56, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Miyata, K.; Oba, M.; Nakanishi, M.; Fukushima, S.; Yamasaki, Y.; Koyama, H.; Nishiyama, N.; Kataoka, K. Polyplexes from poly(aspartamide) bearing 1,2-diaminoethane side chains induce pH-selective, endosomal membrane destabilization with amplified transfection and negligible cytotoxicity. J. Am. Chem. Soc. 2008, 130, 16287–16294. [Google Scholar] [CrossRef] [PubMed]
- Rehman, Z.U.; Hoekstra, D.; Zuhorn, I.S. Mechanism of polyplex- and lipoplex-mediated delivery of nucleic acids: Real-Time visualization of transient membrane destabilization without endosomal lysis. ACS Nano 2013, 7, 3767–3777. [Google Scholar] [CrossRef]
- Chen, Q.; Osada, K.; Ge, Z.; Uchida, S.; Tockary, T.A.; Dirisala, A.; Matsui, A.; Toh, K.; Takeda, K.M.; Liu, X.; et al. Polyplex micelle installing intracellular self-processing functionalities without free catiomers for safe and efficient systemic gene therapy through tumor vasculature targeting. Biomaterials 2017, 113, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Matsumura, Y.; Maeda, H. A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agents Smancs. Cancer Res. 1986, 46, 6387–6392. [Google Scholar] [CrossRef]
- Wilhelm, S.; Tavares, A.J.; Dai, Q.; Ohta, S.; Audet, J.; Dvorak, H.F.; Chan, W.C.W. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 2016, 1, 16014. [Google Scholar] [CrossRef]
- Lee, H.; Shields, A.F.; Siegel, B.A.; Miller, K.D.; Krop, I.; Ma, C.X.; Lorusso, P.M.; Munster, P.N.; Campbell, K.; Gaddy, D.F.; et al. 64Cu-MM-302 positron emission tomography quantifies variability of enhanced permeability and retention of nanoparticles in relation to treatment response in patients with metastatic breast cancer. Clin. Cancer Res. 2017, 23, 4190–4202. [Google Scholar] [CrossRef] [Green Version]
- Golombek, S.K.; May, J.; Theek, B.; Appold, L.; Drude, N.; Kiessling, F.; Lammers, T. Tumor Targeting via EPR: Strategies to Enhance Patient Responses. Adv. Drug Deliv. Rev. 2018, 130, 17–38. [Google Scholar] [CrossRef] [PubMed]
- Van der Meel, R.; Lammers, T.; Hennink, W.E. Cancer nanomedicines: Oversold or underappreciated? Expert Opin. Drug Deliv. 2017, 14, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kenausis, G.L.; Vörös, J.; Elbert, D.L.; Huang, N.; Hofer, R.; Ruiz-Taylor, L.; Textor, M.; Hubbell, J.A.; Spencer, N.D. Poly(L-lysine)-g-poly(ethylene glycol) layers on metal oxide surfaces: Attachment mechanism and effects of polymer architecture on resistance to protein adsorption. J. Phys. Chem. B 2000, 104, 3298–3309. [Google Scholar] [CrossRef]
- Cabral, H.; Miyata, K.; Osada, K.; Kataoka, K. Block copolymer micelles in nanomedicine applications. Chem. Rev. 2018, 118, 6844–6892. [Google Scholar] [CrossRef] [Green Version]
- Dirisala, A.; Uchida, S.; Toh, K.; Li, J.; Osawa, S.; Tockary, T.A.; Liu, X.; Abbasi, S.; Hayashi, K.; Mochida, Y. Transient stealth coating of liver sinusoidal wall by anchoring two-armed PEG for retargeting nanomedicines. Sci. Adv. 2020, 6, eabb8133. [Google Scholar] [CrossRef]
- Cabral, H.; Matsumoto, Y.; Mizuno, K.; Chen, Q.; Murakami, M.; Kimura, M.; Terada, Y.; Kano, M.R.; Miyazono, K.; Uesaka, M.; et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol. 2011, 6, 815–823. [Google Scholar] [CrossRef]
- Oba, M.; Miyata, K.; Osada, K.; Christie, J.R.; Sanjoh, M.; Li, W.; Fukushima, S.; Ishii, T.; Kano, M.R.; Nishiyama, N.; et al. Polyplex micelles prepared from ω-cholesteryl PEG-polycation block copolymers for systemic gene delivery. Biomaterials 2011, 32, 652–663. [Google Scholar] [CrossRef]
- Boeckle, S.; von Gersdorff, K.; van der Piepen, S.; Culmsee, C.; Wagner, E.; Ogris, M. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J. Gene Med. 2004, 6, 1102–1111. [Google Scholar] [CrossRef]
- Fang, J.; Nakamura, H.; Maeda, H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Deliv. Rev. 2011, 63, 136–151. [Google Scholar] [CrossRef]
- Conner, S.D.; Schmid, S.L. Regulated portals of entry into the cell. Nature 2003, 422, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Mintzer, M.A.; Simanek, E.E. Nonviral vectors for gene delivery. Chem. Rev. 2008, 109, 259–302. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-Viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Lächelt, U.; Wagner, E. Nucleic acid therapeutics using polyplexes: A journey of 50 years (and beyond). Chem. Rev. 2015, 115, 11043–11078. [Google Scholar] [CrossRef]
- Lostalé-Seijo, I.; Montenegro, J. Synthetic materials at the forefront of gene delivery. Nat. Rev. Chem. 2018, 2, 258–277. [Google Scholar] [CrossRef]
- Khalil, I.A.; Sato, Y.; Harashima, H. Recent advances in the targeting of systemically administered non-viral gene delivery systems. Expert Opin. Drug Deliv. 2019, 16, 1037–1050. [Google Scholar] [CrossRef]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-Assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef]
- Hahn, J.; Wickham, S.F.J.; Shih, W.M.; Perrault, S.D. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef]
- Kiviaho, J.K.; Linko, V.; Ora, A.; Tiainen, T.; Järvihaavisto, E.; Mikkilä, J.; Tenhu, H.; Nonappa; Kostiainen, M.A. Cationic polymers for DNA origami coating-examining their binding efficiency and tuning the enzymatic reaction rates. Nanoscale 2016, 8, 11674–11680. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.T.; Gray, M.A.; Xuan, S.; Lin, Y.; Byrnes, J.; Nguyen, A.I.; Todorova, N.; Stevens, M.M.; Bertozzi, C.R.; Zuckermann, R.N.; et al. DNA origami protection and molecular interfacing through engineered sequence-defined peptoids. Proc. Natl. Acad. Sci. USA 2020, 117, 6339–6348. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, N.P.; Matthies, M.; Gür, F.N.; Osada, K.; Schmidt, T.L. Block Copolymer Micellization as a Protection Strategy for DNA Origami. Angew. Chem.–Int. Ed. 2017, 56, 5460–5464. [Google Scholar] [CrossRef]
- Ponnuswamy, N.; Bastings, M.M.C.; Nathwani, B.; Ryu, J.H.; Chou, L.Y.T.; Vinther, M.; Li, W.A.; Anastassacos, F.M.; Mooney, D.J.; Shih, W.M. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017, 8, 15654–15662. [Google Scholar] [CrossRef] [PubMed]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-Less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A phase separation model for transcriptional control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sing, C.E.; Perry, S.L. Recent progress in the science of complex coacervation. Soft Matter 2020, 16, 2885–2914. [Google Scholar] [CrossRef] [Green Version]
- Nott, T.J.; Craggs, T.D.; Baldwin, A.J. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat. Chem. 2016, 8, 569–575. [Google Scholar] [CrossRef]
- Schmidt, N.W.; Jin, F.; Lande, R.; Curk, T.; Xian, W.; Lee, C.; Frasca, L.; Frenkel, D.; Dobnikar, J.; Gilliet, M.; et al. Liquid-Crystalline ordering of antimicrobial peptide-DNA complexes controls TLR9 activation. Nat. Mater. 2015, 14, 696–701. [Google Scholar] [CrossRef]
- Lee, E.Y.; Zhang, C.; Di Domizio, J.; Jin, F.; Connell, W.; Hung, M.; Malkoff, N.; Veksler, V.; Gilliet, M.; Ren, P.; et al. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat. Commun. 2019, 10, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lande, R.; Lee, E.Y.; Palazzo, R.; Marinari, B.; Pietraforte, I.; Santos, G.S.; Mattenberger, Y.; Spadaro, F.; Stefanantoni, K.; Iannace, N.; et al. CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-α production in systemic sclerosis. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lerman, L.S. A Transition to a compact form of DNA in polymer solutions. Proc. Nat. Acad. Sci. USA 1971, 68, 1886–1890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K.; Teaff, N.; D’Ambrosia, J. Maturation of the head of bacteriophage T4. V. A possible DNA packaging mechanism: In vitro cleavage of the head proteins and the structure of the core of the polyhead. J. Supramol. Struct. 1974, 2, 276–301. [Google Scholar] [CrossRef]
- Ichiba, Y.; Yoshikawa, K. Single chain observation on collapse transition in giant DNA induced by negatively-charged polymer. Biochem. Biophys. Res. Commun. 1998, 242, 441–445. [Google Scholar] [CrossRef]
- Akabayov, B.; Akabayov, S.R.; Lee, S.J.; Wagner, G.; Richardson, C.C. Impact of macromolecular crowding on DNA replication. Nat. Commun. 2013, 4, 1615–1624. [Google Scholar] [CrossRef] [Green Version]
- Nakano, S.I.; Miyoshi, D.; Sugimoto, N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef]






© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osada, K. Structural Polymorphism of Single pDNA Condensates Elicited by Cationic Block Polyelectrolytes. Polymers 2020, 12, 1603. https://doi.org/10.3390/polym12071603
Osada K. Structural Polymorphism of Single pDNA Condensates Elicited by Cationic Block Polyelectrolytes. Polymers. 2020; 12(7):1603. https://doi.org/10.3390/polym12071603
Chicago/Turabian StyleOsada, Kensuke. 2020. "Structural Polymorphism of Single pDNA Condensates Elicited by Cationic Block Polyelectrolytes" Polymers 12, no. 7: 1603. https://doi.org/10.3390/polym12071603
APA StyleOsada, K. (2020). Structural Polymorphism of Single pDNA Condensates Elicited by Cationic Block Polyelectrolytes. Polymers, 12(7), 1603. https://doi.org/10.3390/polym12071603