Thin Film Plastic Antibody-Based Microplate Assay for Human Serum Albumin Determination
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Synthesis and Screening of HSA-MIP
2.3. Optimization of Template Imprinting
2.4. Selectivity Test
2.5. Thin Film Polymer Characterization
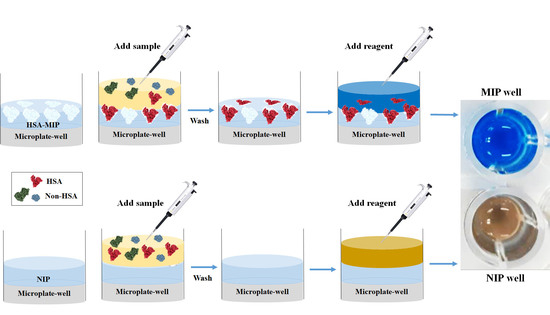
2.6. Polymer-Based Microplate Assay Characterization
3. Results and Discussion
3.1. Optimization of HSA-MIP Synthesis
3.2. Optimization of Template Imprinting
3.3. Thin Film Polymer Characterization
3.4. Selectivity of HSA-MIP
3.5. Polymer-Based Microplate Assay Characterization
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Piletsky, S.A.; Alcock, S.; Turner, A.P. Molecular imprinting: At the edge of the third millennium. Trends Biotechnol. 2001, 19, 9–12. [Google Scholar] [CrossRef]
- Piletsky, S.A.; Turner, N.W.; Laitenberger, P. Molecularly imprinted polymers in clinical diagnostics—Future potential and existing problems. Med. Eng. Phys. 2006, 28, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Bossi, A.; Bonini, F.; Turner, A.P.; Piletsky, S.A. Molecularly imprinted polymers for the recognition of proteins: The state of the art. Biosens. Bioelectron. 2007, 22, 1131–1137. [Google Scholar] [CrossRef]
- Hussain, M.; Wackerlig, J.; Lieberzeit, P.A. Biomimetic strategies for sensing biological species. Biosensors 2013, 3, 89–107. [Google Scholar] [CrossRef] [PubMed]
- Wackerlig, J.; Lieberzeit, P.A. Molecularly imprinted polymer nanoparticles in chemical sensing—Synthesis, characterisation and application. Sens. Actuators B Chem. 2015, 207, 144–157. [Google Scholar] [CrossRef]
- Naseri, M.; Mohammadniaei, M.; Sun, Y.; Ashley, J. The Use of Aptamers and Molecularly Imprinted Polymers in Biosensors for Environmental Monitoring: A Tale of Two Receptors. Chemosensors 2020, 8, 32. [Google Scholar] [CrossRef]
- Zaidi, S.A. Molecular imprinting: A useful approach for drug delivery. Mater. Sci. Energy Technol. 2020, 3, 72–77. [Google Scholar] [CrossRef]
- Culver, H.R.; Peppas, N.A. Protein-Imprinted Polymers: The Shape of Things to Come? Chem. Mater. 2017, 29, 5753–5761. [Google Scholar] [CrossRef]
- Jang, R.; Kim, K.H.; Zaidi, S.A.; Cheong, W.J.; Moon, M.H. Analysis of phospholipids using an open-tubular capillary column with a monolithic layer of molecularly imprinted polymer in capillary electrochromatography-electrospray ionization-tandem mass spectrometry. Electrophoresis 2011, 32, 2167–2173. [Google Scholar] [CrossRef]
- Spizzirri, U.G.; Peppas, N.A. Structural Analysis and Diffusional Behavior of Molecularly Imprinted Polymer Networks for Cholesterol Recognition. Chem. Mater. 2005, 17, 6719–6727. [Google Scholar] [CrossRef]
- Chunta, S.; Suedee, R.; Singsanan, S.; Lieberzeit, P.A. Sensing array based on molecularly imprinted polymers for simultaneous assessment of lipoproteins. Sens. Actuators B Chem. 2019, 298, 126828. [Google Scholar] [CrossRef]
- Malik, A.A.; Nantasenamat, C.; Piacham, T. Molecularly imprinted polymer for human viral pathogen detection. Mater. Sci. Eng. C 2017, 77, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Zhang, Z.; Li, J.; Ma, X.; Chen, L.; Yang, X. Molecular imprinting technology for microorganism analysis. TrAC Trends Anal. Chem. 2018, 106, 190–201. [Google Scholar] [CrossRef]
- Piletsky, S.; Canfarotta, F.; Poma, A.; Bossi, A.M.; Piletsky, S. Molecularly Imprinted Polymers for Cell Recognition. Trends Biotechnol. 2020, 38, 368–387. [Google Scholar] [CrossRef] [PubMed]
- Doumas, B.T.; Watson, W.A.; Biggs, H.G. Albumin standards and the measurement of serum albumin with bromcresol green. Clin. Chim. Acta Int. J. Clin. Chem. 1971, 31, 87–96. [Google Scholar] [CrossRef]
- Lee, P.; Wu, X. Review: Modifications of human serum albumin and their binding effect. Curr. Pharm. Des. 2015, 21, 1862–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stojanovic, Z.; Erdőssy, J.; Keltai, K.; Scheller, F.W.; Gyurcsányi, R.E. Electrosynthesized molecularly imprinted polyscopoletin nanofilms for human serum albumin detection. Anal. Chim. Acta 2017, 977, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, P.G. The measurement of albumin in serum and plasma. Ann. Clin. Biochem. 1985, 22 Pt 6, 565–578. [Google Scholar] [CrossRef]
- Zhang, H. Molecularly Imprinted Nanoparticles for Biomedical Applications. Adv. Mater. 2020, 32, 1806328. [Google Scholar] [CrossRef]
- Oviedo, M.J.; Quester, K.; Hirata, G.A.; Vazquez-Duhalt, R. Determination of conjugated protein on nanoparticles by an adaptation of the Coomassie blue dye method. MethodsX 2019, 6, 2134–2140. [Google Scholar] [CrossRef]
- Lowdon, J.W.; Diliën, H.; Singla, P.; Peeters, M.; Cleij, T.J.; van Grinsven, B.; Eersels, K. MIPs for commercial application in low-cost sensors and assays—An overview of the current status quo. Sens. Actuators B Chem. 2020, 325, 128973. [Google Scholar] [CrossRef] [PubMed]
- Smolinska-Kempisty, K.; Guerreiro, A.; Canfarotta, F.; Cáceres, C.; Whitcombe, M.J.; Piletsky, S. A comparison of the performance of molecularly imprinted polymer nanoparticles for small molecule targets and antibodies in the ELISA format. Sci. Rep. 2016, 6, 37638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moczko, E.; Díaz, R.; Rivas, B.; García, C.; Pereira, E.; Piletsky, S.; Cáceres, C. Molecularly Imprinted Nanoparticles Assay (MINA) in Pseudo ELISA: An Alternative to Detect and Quantify Octopamine in Water and Human Urine Samples. Polymers 2019, 11, 1497. [Google Scholar] [CrossRef] [Green Version]
- Chunta, S.; Suedee, R.; Lieberzeit, P.A. Low-Density Lipoprotein Sensor Based on Molecularly Imprinted Polymer. Anal. Chem. 2016, 88, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Phan, N.V.H.; Sussitz, H.F.; Lieberzeit, P.A. Polymerization Parameters Influencing the QCM Response Characteristics of BSA MIP. Biosensors 2014, 4, 161–171. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, T.X.; Horita, T.J.; Kasravi, B. Understanding and interpreting serum protein electrophoresis. Am. Fam. Physician 2005, 71, 105–112. [Google Scholar] [PubMed]
- Park, Y.M.; Kim, S.J.; Lee, K.J.; Yang, S.S.; Min, B.-H.; Yoon, H.C. Detection of CTX-II in serum and urine to diagnose osteoarthritis by using a fluoro-microbeads guiding chip. Biosens. Bioelectron. 2015, 67, 192–199. [Google Scholar] [CrossRef]
- Nattich-Rak, M.; Dąbkowska, M.; Adamczyk, Z. Microparticle Deposition on Human Serum Albumin Layers: Unraveling Anomalous Adsorption Mechanism. Colloids Interfaces 2020, 4, 51. [Google Scholar] [CrossRef]
- Wright, A.K.; Thompson, M.R. Hydrodynamic structure of bovine serum albumin determined by transient electric birefringence. Biophys. J. 1975, 15, 137–141. [Google Scholar] [CrossRef] [Green Version]
- Tjernberg, A.; Markova, N.; Griffiths, W.J.; Hallén, D. DMSO-related effects in protein characterization. J. Biomol. Screen. 2006, 11, 131–137. [Google Scholar] [CrossRef] [Green Version]
- Majorek, K.A.; Porebski, P.J.; Dayal, A.; Zimmerman, M.D.; Jablonska, K.; Stewart, A.J.; Chruszcz, M.; Minor, W. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol. 2012, 52, 174–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lexa, K.W.; Dolghih, E.; Jacobson, M.P. A structure-based model for predicting serum albumin binding. PLoS ONE 2014, 9, e93323. [Google Scholar] [CrossRef]
- Gibizova, V.V.; Sergeeva, I.A.; Petrova, G.P.; Priezzhev, A.V.; Khlebtsov, N.G. Interaction of albumin and γ-globulin molecules with gold nanoparticles in water solutions. Mosc. Univ. Phys. Bull. 2011, 66, 449–452. [Google Scholar] [CrossRef]
- Chunta, S.; Suedee, R.; Lieberzeit, P.A. High-density lipoprotein sensor based on molecularly imprinted polymer. Anal. Bioanal. Chem. 2018, 410, 875–883. [Google Scholar] [CrossRef] [PubMed]












| Type | MIP before Washing (n = 4) | MIP after Washing (n = 4) | NIP (n = 2) | |||
|---|---|---|---|---|---|---|
| Across (nm) | High (nm) | Across (nm) | Deep (nm) | Across (nm) | Deep (nm) | |
| 12.0 | 0.3 | 12.0 | 0.7 | 50.0 | 2.8 | |
| 22.0 | 0.5 | 17.0 | 0.9 | 43.0 | 1.3 | |
| 14.0 | 0.5 | 27.0 | 1.0 | - | - | |
| 14.0 | 0.3 | 26.0 | 1.5 | - | - | |
| Mean | 15.5 | 0.4 | 20.5 | 1.0 | 46.5 | 2.1 |
| SD | 3.8 | 0.1 | 7.2 | 0.3 | 4.9 | 1.1 |
| Type of Protein | Absorbance | Relative Factor |
|---|---|---|
| HSA | 1.42 ± 0.03 | 1.0 |
| BSA | 1.29 ± 0.01 | 0.9 |
| γ-globulin | 1.01 ± 0.02 | 0.7 |
| α-globulin | 0.75 ± 0.02 | 0.5 |
| β-globulin | 0.59 ± 0.01 | 0.4 |
| Spiked (g/dL) | Measured (n = 3) | Recovery (± SD) (%) | ||
|---|---|---|---|---|
| Mean (g/dL) | SD | %CV | ||
| 2.5 | 2.3 | 0.04 | 2 | 92 ± 2 |
| 5.5 | 5.1 | 0.21 | 4 | 93 ± 4 |
| 7.4 | 7.5 | 0.17 | 2 | 101 ± 2 |
| 8.5 | 8.1 | 0.25 | 3 | 95 ± 3 |
| 8.9 | 9.1 | 0.36 | 4 | 102 ± 4 |
| Type | Concentration of HSA (g/dL) | Mean (g/dL) | SD | %CV |
|---|---|---|---|---|
| Within-run precision (n = 10) | 1 | 0.98 | 0.07 | 7 |
| 10 | 10.16 | 0.37 | 4 | |
| Between-day precision | 1 | 1.01 | 0.09 | 9 |
| (n = 10) | 10 | 10.11 | 0.79 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boonsriwong, W.; Chunta, S.; Thepsimanon, N.; Singsanan, S.; Lieberzeit, P.A. Thin Film Plastic Antibody-Based Microplate Assay for Human Serum Albumin Determination. Polymers 2021, 13, 1763. https://doi.org/10.3390/polym13111763
Boonsriwong W, Chunta S, Thepsimanon N, Singsanan S, Lieberzeit PA. Thin Film Plastic Antibody-Based Microplate Assay for Human Serum Albumin Determination. Polymers. 2021; 13(11):1763. https://doi.org/10.3390/polym13111763
Chicago/Turabian StyleBoonsriwong, Worachote, Suticha Chunta, Nonthawat Thepsimanon, Sanita Singsanan, and Peter A. Lieberzeit. 2021. "Thin Film Plastic Antibody-Based Microplate Assay for Human Serum Albumin Determination" Polymers 13, no. 11: 1763. https://doi.org/10.3390/polym13111763
APA StyleBoonsriwong, W., Chunta, S., Thepsimanon, N., Singsanan, S., & Lieberzeit, P. A. (2021). Thin Film Plastic Antibody-Based Microplate Assay for Human Serum Albumin Determination. Polymers, 13(11), 1763. https://doi.org/10.3390/polym13111763