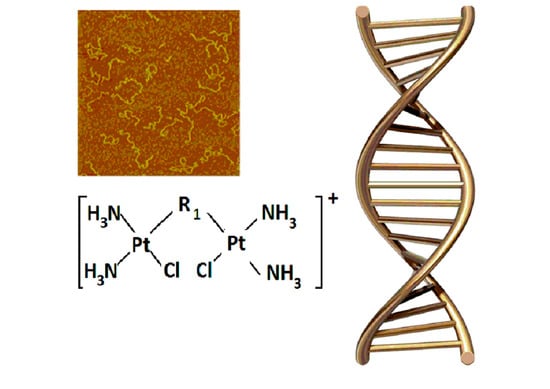
DNA Conformational Changes Induced by Its Interaction with Binuclear Platinum Complexes in Solution Indicate the Molecular Mechanism of Platinum Binding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Low Gradient Viscosity, (LGV)
2.2. Flow Birefringence (FB)
2.3. Spectral Methods
2.4. Atomic Force Microscopy (AFM)
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jain, A. Multifunctional, heterometallic ruthenium-platinum complexes with medicinal applications. Coord. Chem. Rev. 2019, 401, 213067. [Google Scholar] [CrossRef]
- Tong, K.-C.; Wan, P.-K.; Lok, C.-N.; Che, C.-M. Dynamic supramolecular self-assembly of platinum (II) complexes perturbs an autophagy–lysosomal system and triggers cancer cell death. Chem. Sci. 2021, 12, 15229–15238. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; He, W.; Song, Y.; Zhao, J.; Song, Z.; Shan, Y.; Hua, W.; Sun, Y. Multifunctional platinum (IV) complex bearing HDAC inhibitor and biotin moiety exhibiting prominent cytotoxicity and tumor targeting ability. Dalton Trans. 2022, 51, 7343–7351. [Google Scholar] [CrossRef] [PubMed]
- Kasyanenko, N.; Bakulev, V.; Perevyazko, I.; Nekrasova, T.; Nazarova, O.; Slita, A.; Zolotova, Y.; Panarin, E. Model system for multifunctional delivery nanoplatforms based on DNA-Polymer complexes containing silver nanoparticles and fluorescent dye. J. Biotechnol. 2016, 236, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Nie, J.; Wang, D.; Wu, H.; Sun, L.; Wang, X.; Wu, J. Engineering a multi-target therapy nanoplatform against tumor growth and metastasis via a novel NSAID-Pt(iv) prodrug. Chem. Commun. 2022, 58, 3803–3806. [Google Scholar] [CrossRef] [PubMed]
- Shirsath, N.R.; Goswami, A.K. Nanocarriers Based Novel Drug Delivery as Effective Drug Delivery: A. Review. Curr. Nanomater. 2019, 4, 71–83. [Google Scholar] [CrossRef]
- Li, F.; Liang, Y.; Wang, M.; Xu, X.; Zhao, F.; Wang, X.; Sun, Y.; Chen, W. Multifunctional nanoplatforms as cascade-responsive drug-delivery carriers for effective synergistic chemo-photodynamic cancer treatment. J. Nanobiotechnol. 2021, 19, 140. [Google Scholar] [CrossRef]
- Rosenberg, B.; van Camp, L.; Krigas, T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef]
- Rosenberg, B.; van Camp, L.; Trosko, J.E.; Mansour, V.H. Platinum compounds: A new class of potent antitumour agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef]
- Kidani, Y.; Inagaki, K.; Iigo, M.; Hoshi, A.; Kuretani, K. Antitumor activity of 1,2-diaminocyclohexane-platinum complexes against sarcoma-180 ascites form. J. Med. Chem. 1978, 12, 1315–1318. [Google Scholar] [CrossRef]
- Harrap, K.R. Preclinical studies identifying carboplatin as a viable cisplatin alternative. Cancer Treat. Rev. 1985, 12, 21–33. [Google Scholar] [CrossRef]
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The Next Generation of Platinum Drugs: Targeted Pt(II) Agents, Nanoparticle Delivery, and Pt(IV) Prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Westmaas, J.A.; Buda, F.; Reedijk, J. Platinum (II) compounds with chelating ligands based on pyridine and pyrimidine: Synthesis, characterizations, DFT calculations, cytotoxic assays and binding to a DNA model base. J. Inorg. Biochem. 2009, 103, 1278–1287. [Google Scholar] [CrossRef] [PubMed]
- Legin, A.A.; Jakupec, M.A.; Bokach, N.A.; Tyan, M.R.; Kukushkin, V.Y.; Keppler, B.K. Guanidine platinum (II) complexes: Synthesis, in vitro antitumor activity, and DNA interactions. J. Inorg. Biochem. 2014, 133, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Stetsenko, A.I.; Yakovlev, K.I.; D’yachenko, S.A. Platinum (II) Complexes of Purine and Pyrimidine Bases and Their Nucleosides. Russ. Chem. Rev. 1987, 56, 875–893. [Google Scholar] [CrossRef]
- Stetsenko, A.I.; Yakovlev, K.I.; Alekseeva, G.M.; Konovalova, A.L. Triamine-type complexes of platinum (II) and their antitumor properties. Theor. Exp. Chem. 1991, 27, 308–314. [Google Scholar] [CrossRef]
- Ivanov, V.B.; Bystrova, E.I.; Yakovlev, K.I.; Rozhkova, N.D.; Stetsenko, A.I.; Adamov, O.M. Growth inhibiting and cytostatic activities of triamine platinum II complexes with heterocyclic amines. Izv. Ros. Akad. Nauk. Ser. Biol. 1992, 6, 898–907. [Google Scholar]
- Komeda, M.S.; Lin, Y.-L.; Chikuma, M.A. A Tetrazolato-Bridged Dinuclear Platinum (II) Complex Exhibits Markedly High in vivo Antitumor Activity against Pancreatic Cancer. ChemMedChem 2011, 6, 987–990. [Google Scholar] [CrossRef]
- Uemura, M.; Hoshiyama, M.; Furukawa, A.; Sato, T.; Higuchia, Y.; Komeda, S. Highly efficient uptake into cisplatin-resistant cells and the isomerization upon coordinative DNA binding of anticancer tetrazolato-bridged dinuclear platinum (II) complexes. Metallomics 2015, 7, 1488–1496. [Google Scholar] [CrossRef]
- Rajković, S.; Rychlewska, U.; Warzajtis, B.; Ašanina, D.P.; Živković, M.D.; Djuran, M.I. Disparate behavior of pyrazine and pyridazine platinum (II) dimers in the hydrolysis of histidine- and methionine-containing peptides and unique crystal structure of {[Pt(en)Cl]2(μ-pydz)}Cl2 with a pair of NH⋯Cl−⋯HN hydrogen bonds supporting the pyridazine bridge. Polyhedron 2014, 67, 279–285. [Google Scholar]
- Kasyanenko, N.; Aia, E.; Bogdanov, A.; Kosmotynskaya, Y.; Yakovlev, K. Comparison of DNA complexation with antitumor agent cis-DDP and binuclear bivalent platinum compound containing pyrazine. Mol. Biol. 2002, 36, 594–600. [Google Scholar] [CrossRef]
- Berners-Price, S.J.; Davies, M.S.; Cox, J.W.; Thomas, D.S.; Farrell, N. Competitive reactions of interstrand and intrastrand DNA-Pt adducts: A dinuclear-platinum complex preferentially forms a 1,4-interstrand cross-link rather than a 1,2 intrastrand cross-link on binding to a GG 14-mer duplex. Chemistry 2003, 9, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Stetsenko, A.I.; Yakovlev, K.I.; Rozhkova, N.D.; Pogareva, V.G.; Kazakov, S.A. Binuclear Pt (II) complexes with bridging molecules of polyethylene diamine. Coord. Chem. 1990, 16, 560–565. (In Russian) [Google Scholar]
- Yakovlev, K.I.; Rozhkova, N.D.; Stetsenko, A.I. Mono- and binuclear platinum (II) complexes with benzotriazole. J. Inorg. Chem. 1991, 36, 120–127. (In Russian) [Google Scholar]
- Konovalova, A.L.; Yakovlev, K.I.; Stetsenko, A.I.; Rozhkova, N.D.; Gerasimova, G.K.; Ivanova, T.I.; Kamaletdinov, N.S.; Sinditskii, V.P. Antitumor activity of binuclear cationic complexes of platinum (II). Pharm. Chem. J. 1994, 28, 21–24. [Google Scholar] [CrossRef]
- Roberts, J.J.; Thomson, A.J. The mechanism of action of antitumor platinum compounds. Prog. Nucleic Acid Res. Mol. Biol. 1979, 22, 71–133. [Google Scholar]
- Lippert, B. Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug, 1st ed.; Wiley: Weinheim, Germany, 1999. [Google Scholar]
- Watanabe, Y.; Maekawa, M. Methylation of DNA in cancer. Adv. Clin. Chem. 2010, 52, 145–167. [Google Scholar]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Koldobskiy, M.A.; Göndör, A. Epigenetic modulators, modifiers and mediators in cancer etiology and progression. Nat. Rev. Genet. 2016, 17, 284–299. [Google Scholar] [CrossRef]
- Zeller, C.; Dai, W.; Steele, N.L.; Siddiq, A.; Walley, A.J.; Wilhelm-Benartzi, C.S.; Rizzo, S.; van der Zee, A.; Plumb, J.A.; Brown, R. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene 2012, 31, 4567–4576. [Google Scholar] [CrossRef] [Green Version]
- Lund, R.J.; Huhtinen, K.; Salmi, J.; Rantala, J.; Nguyen, E.V.; Moulder, R.; Goodlett, D.R.; Lahesmaa, R.; Carpén, O. DNA methylation and Transcriptome Changes Associated with Cisplatin Resistance in Ovarian Cancer. Sci. Rep. 2017, 7, 1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.W.; Zheng, Y.; Wang, J.Z.; Lu, X.X.; Wang, Z.; Chen, L.B.; Guan, X.X.; Tong, J.D. Integrated analysis of DNA methylation and mRNA expression profiling reveals candidate genes associated with cisplatin resistance in non-small cell lung cancer. Epigenetics 2014, 9, 896–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Byrne, K.J.; Barr, M.P.; Gray, S.G. The Role of Epigenetics in Resistance to Cisplatin Chemotherapy in Lung Cancer. Cancers 2011, 3, 1426–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wermann, H.; Stoop, H.; Gillis, A.J.M.; Honecker, F.; van Gurp, R.; Ammerpohl, O.; Richter, J.; Oosterhuis, J.W.; Bokemeyer, C.; Looijenga, L.H.J. Global DNA methylation in fetal human germ cells and germ cell tumours: Association with differentiation and cisplatin resistance. J. Path. 2010, 221, 433–442. [Google Scholar] [CrossRef]
- Morel, E.; Beauvineau, C.; Naud-Martin, D.; Landras-Guetta, C.; Verga, D.; Ghosh, D.; Achelle, S.; Mahuteau-Betzer, F.; Bombard, S.; Teulade-Fichou, M.-P. Selectivity of Terpyridine Platinum Anticancer Drugs for G-quadruplex DNA. Molecules 2019, 24, 404. [Google Scholar] [CrossRef] [Green Version]
- Trajkovski, M.; Morel, E.; Hamon, F.; Bombard, S.; Teulade-Fichou, M.-P.; Plavec, J. Interactions of Pt-ttpy with G-Quadruplexes Originating from Promoter Region of the c-myc Gene Deciphered by NMR and Gel Electrophoresis Analysis. Chem. Eur. J. 2015, 21, 7798–7807. [Google Scholar] [CrossRef]
- Bertrand, H.; Bombard, S.; Monchaud, D.; Talbot, E.; Guédin, A.; Mergny, J.-L.; Grünert, R.; Bednarskid, P.J.; Teulade-Fichou, M.-P. Exclusive platination of loop adenines in the human telomeric G-quadruplex. Org. Biomol. Chem. 2009, 7, 2864–2871. [Google Scholar] [CrossRef]
- Ishibashi, T.; Lippard, S.J. Telomere loss in cells treated with cisplatin. Proc. Natl. Acad. Sci. USA 1998, 95, 4219–4223. [Google Scholar] [CrossRef] [Green Version]
- Renciuk, D.; Zhou, J.; Beaurepaire, L.; Guedin, A.; Bourdoncle, A.; Mergny, J.L. A FRET-based screening assay for nucleic acid ligands. Methods 2012, 57, 122–128. [Google Scholar] [CrossRef]
- Luo, Y.; Granzhan, A.; Verga, D.; Mergny, J.-L. FRET-MC: A fluorescence melting competition assay for studying G4 structures in vitro. Biopolymers 2021, 112, e23415. [Google Scholar] [CrossRef]
- Boger, D.L.; Fink, B.E.; Brunette, S.R.; Tse, W.C.; Hedrick, M.P. A simple, high-resolution method for establishing DNA binding affinity and sequence selectivity. J. Am. Chem. Soc. 2001, 123, 5878–5891. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.A.; Long, E.C. Fluorescent intercalator displacement analyses of DNA binding by the peptide-derived natural products netropsin, actinomycin, and bleomycin. Bioorg. Med. Chem. 2006, 14, 3481–3490. [Google Scholar] [CrossRef] [PubMed]
- Eigner, J.; Doty, P. The native, denatured and renatured states of deoxyribonucleic acid. J. Mol. Biol. 1965, 12, 549–580. [Google Scholar] [CrossRef]
- Spirin, A.S. Spectrophotometric determination of total nucleic acids. Biochemistry 1958, 23, 656–662. [Google Scholar]
- Kas’ianenko, N.A.; Valueva, S.V.; Smorygo, N.A.; D’iachenko, S.A.; Frisman, E.V. Study of the interaction of a DNA molecule with coordination compounds of divalent platinum I. Effect of cis-diaminodichloroplatinum on molecular parameters of DNA in solution. Mol. Biol. 1995, 29, 345–353. [Google Scholar]
- Kas’ianenko, N.A.; Karymov, M.A.; D’iachenko, S.A.; Smorygo, N.A.; Frisman, E.V. Interaction of DNA molecules with divalent platinum coordination complexes. II. Effect of the nature and location of ligands in the first platinum coordination sphere. Mol. Biol. 1995, 29, 585–596. [Google Scholar]
- Kasyanenko, N.A.; Prokhorova, S.A.; Haya Enriquez, E.F.; Sudakova, S.S.; Frisman, E.V.; Dyachenko, S.A.; Smorygo, N.A.; Ivin, B.A. Interaction of protonated DNA with trans-dichlorodiammineplatinum (II). Colloids Surf. A Physicochem. Eng. Asp. 1999, 148, 121–128. [Google Scholar] [CrossRef]
- Tsvetkov, V.N.; Eskin, V.E.; Frenkel, S.Y. Structure of Macromolecules in Solutions; Nauka: Moscow, Russia, 1964. [Google Scholar]
- Frisman, E.V.; Kas’ianenko, N.A. Hydrodynamic and optical behavior of DNA molecule in the range of high ionic strength. Mol. Biol. 1990, 24, 318–327. [Google Scholar]
- Thomson, A.J. The mechanism of action of anti-tumour platinum compounds. Platin. Met. Rev. 1977, 21, 2–15. [Google Scholar]
- Fichtinger-Schepman, A.M.; Lohman, P.H.; Reedijk, J. Detection and quantification of adducts formed upon interaction of diamminedichloroplatinum (II) with DNA, by anion-exchange chromatography after enzymatic degradation. Nucleic Acids Res. 1982, 10, 5345–5356. [Google Scholar] [CrossRef] [Green Version]
- Reed, E.; Yuspa, S.H.; Zwelling, L.A.; Ozols, R.F.; Poirier, M.C. Quantitation of cisplatin-DNA intrastrand adducts in testicular and ovarian cancer patients receiving cisplatin chemotherapy. J. Clin. Investig. 1986, 77, 545–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwelling, L.A.; Anderson, T.; Kohn, K.W. DNA-protein and DNA interstrand cross-linking by cis- and trans-platinum (II) diamminedichloride in L1210 mouse leukemia cells and relation to cytotoxicity. Cancer Res. 1979, 39, 365–369. [Google Scholar] [PubMed]
- Huang, H.; Woo, J.; Alley, S.C.; Hopkins, P.B. DNA-DNA interstrand cross-linking by cis- diamminedichloroplatinum (II): N7(dG)-to-N7(dG) cross-linking at 5’-d(GC) in synthetic oligonucleotides. Bioorg. Med. Chem. 1995, 3, 659–669. [Google Scholar] [CrossRef]
- Noll, D.M.; Mason, T.M.; Miller, P.S. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 2006, 106, 277–301. [Google Scholar] [CrossRef] [PubMed]
- Clement, R.M.; Sturm, J.; Daune, M.P. Interaction of metallic cations with DNA VI. Specific binding of Mg++ and Mn++. Biopolymers 1973, 12, 405–421. [Google Scholar] [CrossRef]
- Kas’ianenko, N.A.; D’iakonova, N.E.; Frisman, E.V. A study of the molecular mechanism of DNA interaction with divalent metal ions. Mol. Biol. 1989, 23, 975–982. [Google Scholar]
- Horácek, P.; Drobník, J. Interaction of cis-dichlorodiammineplatinum (II) with DNA. Biochim. Biophys. Acta 1971, 254, 341–347. [Google Scholar] [CrossRef]
- Macquet, J.P.; Butour, J.L. A cirular dichroism study of DNA-platinum complexes. Differentiation between monofunctional, cis-bidentate and trans-bidentate platinum fixation on a series of DNA. Eur. J. Biochem. 1978, 83, 375–387. [Google Scholar] [CrossRef]
- Akimenko, N.; Cheltsov, P.; Balcarová, Z.; Kleinwächter, V.; Yevdokimov, Y. Study of interactions of platinum (II) compounds with DNA by means of CD spectra of solutions and liquid crystalline microphases of DNA. Gen. Physiol. Biophys. 1985, 4, 597–608. [Google Scholar]
- Kasyanenko, N.; Arikainen, N.; Frisman, E. Investigation of DNA complexes with iron ions in solution. Biophys. Chem. 1998, 70, 93–100. [Google Scholar] [CrossRef]
- Kasyanenko, N.A.; Zanina, A.V.; Nazarova, O.V.; Panarin, E.F. DNA Interaction with Complex Ions in Solution. Langmuir 1999, 15, 7912–7917. [Google Scholar] [CrossRef]
- Kas’yanenko, N.A.; Abramchuk, S.S.; Blagodatskikh, I.V.; Bogdanov, A.A.; Gallyamov, M.O.; Kononov, A.I.; Kosmotynskaya, Y.V.; Sitnikova, N.L.; Khokhlov, A.R. Study of DNA complexation with platinum coordination compounds. Polym. Sci. –Ser. A 2003, 45, 960–968. [Google Scholar]
- Serebryanskaya, T.V.; Kinzhalov, M.A.; Bakulev, V.; Alekseev, G.; Andreeva, A.; Gushchin, P.V.; Protas, A.V.; Smirnov, A.S.; Panikorovskii, T.L.; Lippmann, P.; et al. Water soluble palladium (II) and platinum (II) acyclic diaminocarbene complexes: Solution behavior, DNA binding, and antiproliferative activity. New J. Chem. 2020, 44, 5762–5773. [Google Scholar] [CrossRef]
- Protas, A.V.; Popova, E.A.; Mikolaichuk, O.V.; Porozov, Y.B.; Mehtiev, A.R.; Ott, I.; Alekseev, G.V.; Kasyanenko, N.A.; Trifonov, R.E. Synthesis, DNA and BSA binding of Pd (II) and Pt (II) complexes featuring tetrazolylacetic acids and their esters. Inorg. Chim. Acta 2018, 473, 133–144. [Google Scholar]
- Manzini, G.; Barcellona, M.L.; Avitabile, M.; Quadrifoglio, F. Interaction of diamidino-2-phenylindole (DAPI) with natural and synthetic nucleic acids. Nucleic Acids Res. 1983, 11, 8861–8876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubista, M.; Aakerman, B.; Norden, B. Characterization of interaction between DNA and 4′,6-diamidino-2-phenylindole by optical spectroscopy. Biochemistry 1987, 26, 4545–4553. [Google Scholar] [CrossRef] [PubMed]
- Luck, G.; Zimmer, C.; Snatzke, G. Circular dichroism of protonated DNA. Biochim. Biophys. Acta 1968, 169, 548–549. [Google Scholar] [CrossRef]
- Kasyanenko, N.A.; Bartoshevich, S.F.; Frisman, E.V. Study of influence of pH media on conformation of DNA molecule. Mol. Biol. 1985, 19, 1386–1393. [Google Scholar]












Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasyanenko, N.; Qiushi, Z.; Bakulev, V.; Sokolov, P.; Yakovlev, K. DNA Conformational Changes Induced by Its Interaction with Binuclear Platinum Complexes in Solution Indicate the Molecular Mechanism of Platinum Binding. Polymers 2022, 14, 2044. https://doi.org/10.3390/polym14102044
Kasyanenko N, Qiushi Z, Bakulev V, Sokolov P, Yakovlev K. DNA Conformational Changes Induced by Its Interaction with Binuclear Platinum Complexes in Solution Indicate the Molecular Mechanism of Platinum Binding. Polymers. 2022; 14(10):2044. https://doi.org/10.3390/polym14102044
Chicago/Turabian StyleKasyanenko, Nina, Zhang Qiushi, Vladimir Bakulev, Petr Sokolov, and Konstantin Yakovlev. 2022. "DNA Conformational Changes Induced by Its Interaction with Binuclear Platinum Complexes in Solution Indicate the Molecular Mechanism of Platinum Binding" Polymers 14, no. 10: 2044. https://doi.org/10.3390/polym14102044
APA StyleKasyanenko, N., Qiushi, Z., Bakulev, V., Sokolov, P., & Yakovlev, K. (2022). DNA Conformational Changes Induced by Its Interaction with Binuclear Platinum Complexes in Solution Indicate the Molecular Mechanism of Platinum Binding. Polymers, 14(10), 2044. https://doi.org/10.3390/polym14102044