Oral Microbiome in Orthodontic Acrylic Retainer
Abstract
:1. Introduction
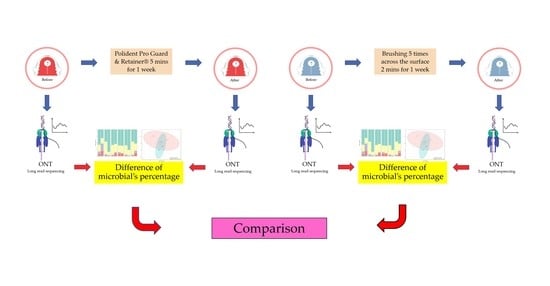
2. Materials and Methods
2.1. Study Design and Participants
- Wear the acrylic retainer for 24 h.
- Wear the acrylic retainer for more than 6 months, except when eating.
- Nonsmoker.
- Without any medication that reduces saliva flow.
- All acrylic retainers are fabricated in the same laboratory.
- Use of steroid-based or antibacterial mouthwash or broad-spectrum antibiotics for 2 weeks before the study.
2.2. Cleaning the Acrylic Retainers
2.3. Plaque Collection
2.4. DNA Extraction
2.5. DNA Preparation and 16S Microbiome Sequencing
2.6. Data and Statistical Analysis
3. Results
3.1. Demographic and Clinical Information
3.2. Phylum, Genus and Species Composition of Biofilms adhering to the Acrylic Retainers
3.3. Diversity of Bacterial Composition on Biofilms of the Acrylic Retainers
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eroglu, A.K.; Baka, Z.M.; Arslan, U. Comparative evaluation of salivary microbial levels and periodontal status of patients wearing fixed and removable orthodontic retainers. Am. J. Orthod. Dentofac. Orthop. 2019, 156, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, K.C.; Pattanaik, S. Modified Wrap-Around Retainer: A Quick Tip To Enhance the Retention of the Appliance. J. Clin. Diagn. Res. 2016, 10, ZH01. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, I.T.; Enoki, C.; Ito, I.Y.; Matsumoto, M.A.; Nelson-Filho, P. Evaluation of home disinfection protocols for acrylic baseplates of removable orthodontic appliances: A randomized clinical investigation. Am. J. Orthod. Dentofac. Orthop. 2011, 140, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Shawesh, M.; Bhatti, B.; Usmani, T.; Mandall, N. Hawley retainers full- or part-time? A randomized clinical trial. Eur. J. Orthod. 2010, 32, 165–170. [Google Scholar] [CrossRef]
- Lucchese, A.; Bondemark, L.; Marcolina, M.; Manuelli, M. Changes in oral microbiota due to orthodontic appliances: A systematic review. J. Oral Microbiol. 2018, 10, 1476645. [Google Scholar] [CrossRef] [PubMed]
- Topaloglu-Ak, A.; Ertugrul, F.; Eden, E.; Ates, M.; Bulut, H. Effect of orthodontic appliances on oral microbiota—6 Month follow-up. J. Clin. Pediatr. Dent. 2011, 35, 433–436. [Google Scholar] [CrossRef]
- Moshiri, M.; Eckhart, J.E.; McShane, P.; German, D.S. Consequences of poor oral hygiene during aligner therapy. J. Clin. Orthod. 2013, 47, 494–498. [Google Scholar]
- Dogramaci, E.J.; Littlewood, S.J. Removable orthodontic retainers: Practical considerations. Br. Dent. J. 2021, 230, 723–730. [Google Scholar] [CrossRef]
- Shi, B.; Wu, T.; McLean, J.; Edlund, A.; Young, Y.; He, X.; Lv, H.; Zhou, X.; Shi, W.; Li, H.; et al. The Denture-Associated Oral Microbiome in Health and Stomatitis. mSphere 2016, 1, e00215-16. [Google Scholar] [CrossRef]
- Paranhos, H.F.; Silva-Lovato, C.H.; Souza, R.F.; Cruz, P.C.; Freitas, K.M.; Peracini, A. Effects of mechanical and chemical methods on denture biofilm accumulation. J. Oral Rehabil. 2007, 34, 606–612. [Google Scholar] [CrossRef]
- Gong, S.Q.; Epasinghe, J.; Rueggeberg, F.A.; Niu, L.N.; Mettenberg, D.; Yiu, C.K.; Blizzard, J.D.; Wu, C.D.; Mao, J.; Drisko, C.L.; et al. An ORMOSIL-containing orthodontic acrylic resin with concomitant improvements in antimicrobial and fracture toughness properties. PLoS ONE 2012, 7, e42355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.S.; Lee, J.H.; Kim, S.O.; Song, J.S.; Kim, B.I.; Kim, Y.J.; Lee, J.H. Comparison of the oral microbiome of siblings using next-generation sequencing: A pilot study. Oral Dis. 2016, 22, 549–556. [Google Scholar] [CrossRef]
- Albanna, H.; Farawanah, H.; Aldrees, A. Microbial evaluation of the effectiveness of different methods for cleansing clear orthodontic retainers: A randomized clinical trial. Angle Orthod. 2017, 87, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Sherry, L.; Lappin, G.; O’Donnell, L.E.; Millhouse, E.; Millington, O.R.; Bradshaw, D.J.; Axe, A.S.; Williams, C.; Nile, C.J.; Ramage, G. Viable Compositional Analysis of an Eleven Species Oral Polymicrobial Biofilm. Front Microbiol. 2016, 7, 912. [Google Scholar] [CrossRef]
- Carda-Diéguez, M.; Bravo-González, L.A.; Morata, I.M.; Vicente, A.; Mira, A. High-throughput DNA sequencing of microbiota at interproximal sites. J. Oral Microbiol. 2020, 12, 1687397. [Google Scholar] [CrossRef]
- Matsuo, Y.; Komiya, S.; Yasumizu, Y.; Yasuoka, Y.; Mizushima, K.; Takagi, T.; Kryukov, K.; Fukuda, A.; Morimoto, Y.; Naito, Y.; et al. Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinION™ nanopore sequencing confers species-level resolution. BMC Microbiol. 2021, 21, 35. [Google Scholar] [CrossRef]
- Lanfear, R.; Schalamun, M.; Kainer, D.; Wang, W.; Schwessinger, B. MinIONQC: Fast and simple quality control for MinION sequencing data. Bioinformatics 2019, 35, 523–525. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Perez, H.; Ciuffreda, L.; Flores, C. NanoCLUST: A species-level analysis of 16S rRNA nanopore sequencing data. Bioinformatics 2021, 37, 1600–1601. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Kiatwarawut, K.; Rokaya, D.; Sirisoontorn, I. Antimicrobial Activity of Various Disinfectants to Clean Thermoplastic Polymeric Appliances in Orthodontics. Polymers 2022, 14, 2256. [Google Scholar] [CrossRef]
- Winand, R.; Bogaerts, B.; Hoffman, S.; Lefevre, L.; Delvoye, M.; Van Braekel, J.; Fu, Q.; Roosens, N.H.; De Keersmaecker, S.C.; Vanneste, K. Targeting the 16S rRNA Gene for Bacterial Identification in Complex Mixed Samples: Comparative Evaluation of Second (Illumina) and Third (Oxford Nanopore Technologies) Generation Sequencing Technologies. Int. J. Mol. Sci. 2020, 21, 298. [Google Scholar] [CrossRef] [Green Version]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef]
- Babin, P.J.; Vernier, J.M. Plasma lipoproteins in fish. J. Lipid Res. 1989, 30, 467–489. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.S.; Al-Awadi, S.; Ready, D.; Noar, J. An assessment of the effectiveness of mechanical and chemical cleaning of Essix orthodontic retainer. J. Orthod. 2014, 41, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.S.; Ahmed, N. Nanoindentation and surface roughness profilometry of poly methyl methacrylate denture base materials. Technol. Health Care 2014, 22, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.S. Prosthodontic Applications of Polymethyl Methacrylate (PMMA): An Update. Polymers 2020, 12, 2299. [Google Scholar] [CrossRef]
- Delaney, C.; O’Donnell, L.E.; Kean, R.; Sherry, L.; Brown, J.L.; Calvert, G.; Nile, C.J.; Cross, L.; Bradshaw, D.J.; Brandt, B.W.; et al. Interkingdom interactions on the denture surface: Implications for oral hygiene. Biofilm 2019, 1, 100002. [Google Scholar] [CrossRef]
- Lucena-Ferreira, S.C.; Cavalcanti, I.M.; Cury, A.A. Efficacy of denture cleansers in reducing microbial counts from removable partial dentures: A short-term clinical evaluation. Braz. Dent. J. 2013, 24, 353–356. [Google Scholar] [CrossRef]
- Zhu, B.; Macleod, L.C.; Kitten, T.; Xu, P. Streptococcus sanguinis biofilm formation & interaction with oral pathogens. Future Microbiol. 2018, 13, 915–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Wang, M.; Shen, Y.; Zhang, L.; Pan, Y.; Sun, Y.; Zhang, K. Regulatory Effect of Irresistin-16 on Competitive Dual-Species Biofilms Composed of Streptococcus mutans and Streptococcus sanguinis. Pathogens 2022, 11, 70. [Google Scholar] [CrossRef] [PubMed]
- Treerat, P.; Redanz, U.; Redanz, S.; Giacaman, R.A.; Merritt, J.; Kreth, J. Synergism between Corynebacterium and Streptococcus sanguinis reveals new interactions between oral commensals. ISME J. 2020, 14, 1154–1169. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, J.; Wang, Q.; Fitzsimonds, Z.R.; Miller, D.P.; Sztukowska, M.N.; Jung, Y.J.; Hayashi, M.; Whiteley, M.; Lamont, R.J. Streptococcus gordonii programs epithelial cells to resist ZEB2 induction by Porphyromonas gingivalis. Proc. Natl. Acad. Sci. USA 2019, 116, 8544–8553. [Google Scholar] [CrossRef] [Green Version]







| ID | Age | Gender | Probing Depth | Periodontal Index (PI) | Firmicutes/ Bacteroidetes (F/B) | Body Mass Index (BMI) |
|---|---|---|---|---|---|---|
| 1 | 30 | F | ≤3 | 0.83 | 5.51 | 19.10 |
| 2 | 33 | M | ≤3 | 0.5 | 7.11 | 18.73 |
| 3 | 59 | F | ≤3 | 0.67 | 16.57 | 22.66 |
| 4 | 40 | F | ≤3 | 0.75 | 9.31 | 19.53 |
| 5 | 33 | M | ≤3 | 0.5 | 11.89 | 18.75 |
| 6 | 58 | F | ≤3 | 0.67 | 8.56 | 22.27 |
| Phyla | Chemical Tablet Group (Before) | Chemical Tablet Group (After) | Brushing Group (Before) | Brushing Group (After) | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | Median | Mean | Median | Mean | Median | Mean | Median | |
| Actinobacteria | 2.73% | 2.11% | 7.51% | 9.90% | 3.53% | 1.63% | 7.11% | 9.23% |
| Bacteroidetes | 6.18% | 6.14% | 13.62% | 4.53% | 5.33% | 4.99% | 4.88% | 3.66% |
| Firmicutes | 50.17% | 49.25% | 57.11% | 56.45% | 52.62% | 54.38% | 60.97% | 55.96% |
| Proteobacteria | 40.42% | 36.55% | 24.84% | 16.61% | 38.50% | 34.77% | 26.62% | 26.18% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasibut, P.; Kuvatanasuchati, J.; Thaweboon, B.; Sirisoontorn, I. Oral Microbiome in Orthodontic Acrylic Retainer. Polymers 2022, 14, 3583. https://doi.org/10.3390/polym14173583
Kasibut P, Kuvatanasuchati J, Thaweboon B, Sirisoontorn I. Oral Microbiome in Orthodontic Acrylic Retainer. Polymers. 2022; 14(17):3583. https://doi.org/10.3390/polym14173583
Chicago/Turabian StyleKasibut, Punnisa, Jintakorn Kuvatanasuchati, Boonyanit Thaweboon, and Irin Sirisoontorn. 2022. "Oral Microbiome in Orthodontic Acrylic Retainer" Polymers 14, no. 17: 3583. https://doi.org/10.3390/polym14173583
APA StyleKasibut, P., Kuvatanasuchati, J., Thaweboon, B., & Sirisoontorn, I. (2022). Oral Microbiome in Orthodontic Acrylic Retainer. Polymers, 14(17), 3583. https://doi.org/10.3390/polym14173583