Optimization of Polystyrene Biodegradation by Bacillus cereus and Pseudomonas alcaligenes Using Full Factorial Design
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microplastics Preparation
2.2. Bacterial Cultivation
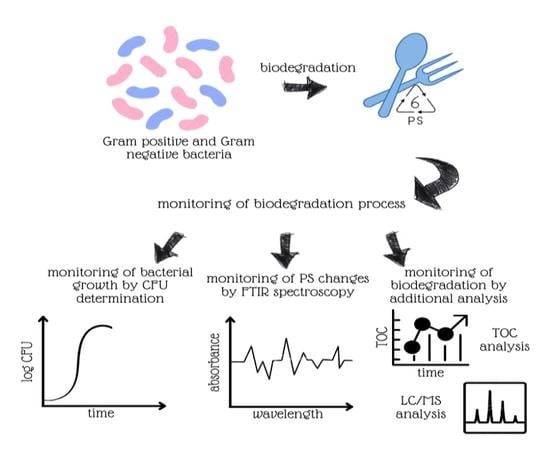
2.3. Biodegradation Experiments
2.3.1. Preliminary Experiments
2.3.2. Main Experiments
2.4. Response Surface Modeling
3. Results and Discussion
3.1. Preliminary Experiment
3.2. Main Experiment
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Plastics—The Facts 2021: An Analysis of European Plastics Production, Demand and Waste Data. Available online: https://plasticseurope.org/wp-content/uploads/2021/12/Plastics-the-Facts-2021-web-final.pdf (accessed on 4 January 2021).
- Bule, K.; Zadro, K.; Tolić, A.; Radin, E.; Miloloža, M.; Ocelić Bulatović, V.; Kučić Grgić, D. Mikroplastika u morskom okolišu Jadrana. Chem. Ind. 2020, 69, 303–310. [Google Scholar] [CrossRef]
- Tareen, A.; Seed, S.; Iqbal, A.; Batool, R.; Jamil, N. Biodeterioration of microplastics: A promising step towards plastics waste management. Polymers 2022, 14, 2275. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.T.; Roberts, T.K.; Lucas, S. An overview on biodegradation of polystyrene and modified polystyrene: The microbial approach. Crit. Rev. Biotechnol. 2018, 38, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Asmita, A.; Shubhamsingh, T.; Tejashree, S. Isolation of plastic degrading microorganisms from soil samples collected at various locations in Mumbai, India. Int. Res. J. Environ. Sci. 2015, 4, 77–85. [Google Scholar]
- Johnston, B.; Radecka, I.; Hill, D.; Chiellini, E.; Ivanova Ilieva, I.; Sikorska, W.; Musioł, M.; Zięba, M.; Marek, A.A.; Keddie, D.; et al. The microbial production of polyhydroxyalkanoates from waste polystyrene fragments attained using oxidative degradation. Polymers 2018, 10, 957. [Google Scholar] [CrossRef] [Green Version]
- Biron, M. Material Selection for Thermoplastic Parts Practical and Advanced Information for Plastics Engineers; Elsevier: Oxford, UK, 2016. [Google Scholar] [CrossRef]
- Yuan, W.-J.; Zhao, W.; Wu, G.; Zhao, H.-B. A phosphorus-nitrogen-carbon synergistic nanolayered flame retardant for polystyrene. Polymers 2022, 14, 2055. [Google Scholar] [CrossRef] [PubMed]
- Hahladakis, J.N.; Velis, C.A.; Weber, R.; Iacovvidou, E.; Purnell, P. An overview of chemical additives present in plastics: Migration, release, fate and environmental impact during their use, disposal and recycling. J. Hazard. Mater. 2018, 344, 179–199. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; He, C.; Han, W.; Song, J.; Li, H.; Zhang, Y.; Jing, X.; Wu, W. Exposure pathways, levels and toxicity of polybrominated diphenyl ethers in humans: A review. Environ. Res. 2020, 187, 109531. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Osako, M.; Sakai, S.-I. Leaching characteristics of polybrominated diphenyl ethers (PBDEs) from flame-retardant plastics. Chemosphere 2006, 65, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Kim, S.-K.; Hyun, J.-H. Environmental distribution of styrene oligomers (SOs) coupled with their source characteristics: Tracing the origin of SOs in the environment. J. Hazard. Mater. 2020, 398, 122968. [Google Scholar] [CrossRef] [PubMed]
- Tawfik, M.S.; Huyghebaert, A. Polystyrene cups and containers: Styrene migration. Food Addit. Contam. 1998, 15, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Auta, H.S.; Emenike, C.U.; Fauziah, S.H. Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environ. Pollut. 2017, 231, 1552–1559. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, N.; Montazer, Z.; Sharma, P.K.; Levin, D.B. Microbial and enzymatic degradation of synthetic plastics. Front. Microbiol. 2020, 11, 580709. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Lee, H.M.; Yu, H.C.; Jeon, E.; Lee, S.; Li, J.; Kim, D.H. Biodegradation of polystyrene by Pseudomonas sp. isolated from the gut of superworms (larvae of Zophobas atratus). Environ. Sci. Technol. 2020, 54, 6987–6996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pedersen, J.N.; Eser, B.E.; Guo, Z. Biodegradation of polyethylene and polystyrene: From microbial deterioration to enzyme discovery. Biotechnol. Adv. 2022, 60, 107991. [Google Scholar] [CrossRef] [PubMed]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques-classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 2016, 32, 1461–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kale, S.K.; Deshmukh, A.G.; Dudhare, M.S.; Patil, V.B. Microbial degradation of plastic: A review. J. Biochem. Technol. 2015, 6, 952–961. [Google Scholar]
- Yuan, J.; Ma, J.; Sun, Y.; Zhou, T.; Zhao, Y.; Yu, F. Microbial degradation and other environmental aspects of microplastics/plastics. Sci. Total Environ. 2020, 715, 136968. [Google Scholar] [CrossRef]
- Tischler, D.; Kaschabek, S.R. Microbial Styrene Degradation: From Basics to Biotechnology. In Microbial Degradation of Xenobiotics; Singh, S.N., Ed.; Chapter 3; Springer: Berlin/Heidelberg, Germany, 2012; pp. 67–99. [Google Scholar] [CrossRef]
- Nakamiya, K.; Sakasita, G.; Ooi, T.; Kinoshita, S. Enzymic degradation of polystyrene by hydroquinone peroxidase of Azotobacter beijerinckii HM121. J. Ferment. Bioeng. 1997, 84, 480–482. [Google Scholar] [CrossRef]
- Hou, L.; Majumder, E.L. Potential for and distribution of enzymatic biodegradation of polystyrene by environmental microorganisms. Materials 2021, 14, 503. [Google Scholar] [CrossRef]
- Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Plastic biodegradation: Frontline microbes and their enzymes. Sci. Total Environ. 2021, 759, 143536. [Google Scholar] [CrossRef]
- Ali, S.S.; Elsamahy, T.; Al-Tohamy, R.; Zhu, D.; Mahmoud, Y.A.; Koutra, E.; Metwally, M.A.; Kornaros, M.; Sun, J. Plastic wastes biodegradation: Mechanisms, challenges and future prospects. Sci. Total Environ. 2021, 780, 146590. [Google Scholar] [CrossRef]
- Mohan, A.J.; Sekhar, V.C.; Bhaskar, T.; Madhavan Nampoothiri, K. Microbial assisted high impact polystyrene (HIPS) degradation. Bioresour. Technol. 2016, 213, 204–207. [Google Scholar] [CrossRef]
- Kučić Grgić, D.; Miloloža, M.; Lovrinčić, E.; Kovačević, A.; Cvetnić, M.; Ocelić Bulatović, V.; Prevarić, V.; Bule, K.; Ukić, Š.; Markić, M.; et al. Bioremediation of MP-polluted waters using bacteria Bacillus licheniformis, Lysinibacillus massiliensis, and mixed culture of Bacillus sp. and Delftia acidovorans. Chem. Biochem. Eng. Q. 2021, 35, 205–224. [Google Scholar] [CrossRef]
- Atiq, N.; Ahmed, S.; Ishtiaq, A.M.; Andleeb, S.; Ahmad, B.; Robson, G. Isolation and identification of polystyrene biodegrading bacteria from soil. Afr. J. Microbiol. Res. 2010, 4, 1537–1541. Available online: http://www.academicjournals.org/ajmr (accessed on 17 July 2022).
- Kundu, D.; Hazra, C.; Chatterjee, A.; Chaudhari, A.; Mishra, S. Biopolymer and biosurfactant-graft-calcium sulfate/polystyrene nanocomposites: Thermophysical, mechanical and biodegradation studies. Polym. Deg. Stab. 2014, 107, 37–52. [Google Scholar] [CrossRef]
- Mor, R.; Sivan, A. Biofilm formation and partial biodegradation of polystyrene by the actinomycete Rhodococcus ruber. Biodegradation 2008, 19, 851–858. [Google Scholar] [CrossRef]
- Motta, O.; Proto, A.; De Carlo, F.; De Caro, F.; Santoro, E.; Brunetti, L.; Capunzo, M. Utilization of chemically oxidized polystyrene as co-substrate by filamentous fungi. Int. J. Hyg. Environ. Health 2009, 212, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.G.; Hinduja, M.; Sujitha, K.; Rajan, N.N.; Dharani, G. Biodegradation of polystyrene by deep-sea Bacillus paralicheniformis G1 and genome analysis. Sci. Total Environ. 2021, 774, 145002. [Google Scholar] [CrossRef]
- Kim, H.-W.; Jo, J.H.; Kim, Y.-B.; Le, T.-K.; Cho, C.-W.; Yun, C.-H.; Chi, W.S.; Yeom, S.-J. Biodegradation of polystyrene by bacteria from the soil in common environments. J. Hazard. Mater. 2021, 416, 126239. [Google Scholar] [CrossRef]
- Shabbir, S.; Faheem, M.; Ali, N.; Kerr, P.G.; Wang, L.-F.; Kuppusamy, S.; Li, Y. Perriphytic biofilm: An innovative approach for biodegradation of microplastics. Sci. Total Environ. 2020, 717, 137064. [Google Scholar] [CrossRef]
- Rao, R.S.; Kumar, C.G.; Prakasham, R.S.; Hobbs, P.J. The Taguchi methodology as a statistical tool for biotechnological applications: A critical appraisal. Biotechnol. J. 2008, 3, 510–523. [Google Scholar] [CrossRef]
- Joutey, N.T.; Bahafid, W.; Sayel, H.; Maataoui, H.; Errachidi, F.; Ghachtouli, N.E. Use of experimental factorial design for optimization of hexavalent chromium removal by a bacterial consortium: Soil microcosm bioremediation. Soil Sediment Contam. 2014, 24, 129–142. [Google Scholar] [CrossRef]
- Anthony, J. Full Factorial designs. In Design of Experiments for Engineers and Scientists, 2nd ed.; Anthony, J., Ed.; Elsevier: Oxford, MS, USA, 2014; Chapter 6; pp. 63–85. [Google Scholar] [CrossRef]
- Ravanipour, M.; Kalantary, R.R.; Mohseni-Bandpi, A.; Esrafili, A.; Farzadkia, M.; Hashemi-Najafabadi, S. Experimental design approach to the optimization of PAHs bioremediation from artificially contaminated soil: Application of variables screening development. J. Environ. Health 2015, 13, 22. [Google Scholar] [CrossRef] [Green Version]
- Briški, F.; Kopčić, N.; Ćosić, I.; Kučić, D.; Vuković, M. Biodegradation of tobacco waste by composting: Genetic identification of nicotine-degrading bacteria and kinetic analysis of transformations in leachate. Chem. Pap. 2012, 66, 1103–1110. [Google Scholar] [CrossRef]
- Benson, H.J. Microbiological Applications, A Laboratory Manual in General Microbiology, 8th ed.; The McGraw-Hill Companies: New York, NY, USA, 2001; pp. 154–156, 160, 168, 169. [Google Scholar]
- Kyaw, B.M.; Champakalakshmi, R.; Kishore Sakharkar, M.; Lim, C.S.; Sakharkar, K.R. Biodegradation of low density polyethene (LDPE) by Pseudomonas species. Indian J. Microbiol. 2012, 52, 411–419. [Google Scholar] [CrossRef] [Green Version]
- Black, J.G. Microbiology: Principles and Explorations, 8th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2012; p. 975. [Google Scholar]
- ISO 11348-3; Water Quality—Determination of the Inhibitory Effect of Water Samples on the Light Emission of Vibrio fischeri (Luminescent bacteria Test)—Part 3: Method Using Freeze-Dried Bacteria. ISO Publishing: Geneva, Switzerland, 2007.
- Schmidt-Rohr, K. Oxygen is the high-energy molecule powering complex multicellular life: Fundamental corrections to traditional bioenergetics. ACS Omega 2020, 5, 2221–2233. [Google Scholar] [CrossRef]
- Price, D.; Horrocks, A.R. Combustion processes of textile fibres. In Handbook of Fire Resistant Textiles; Woodhead Publishing Limited: Cambridge, UK, 2013; pp. 3–25. [Google Scholar] [CrossRef]
- El-Arabi, T.F.; Griffiths, M.W. Bacillus cereus. In Foodborne Infections and Intoxications, 4th ed.; Morris, J.G., Morris, E., Eds.; Elsevier: Oxford, MS, USA, 2013; Chapter 29; pp. 401–407. [Google Scholar] [CrossRef]
- Yuan, Z.; Nag, R.; Cummins, E. Human health concerns regarding microplastics in the aquatic environment-From marine to food systems. Sci. Total Environ. 2022, 823, 153730. [Google Scholar] [CrossRef]
- Čačko, S.; Pančić, E.; Zokić, I.; Miloloža, M.; Kučić Grgić, D. Aditivi u plastici—Potencijalno štetni učinci na ekosustav. Chem. Ind. 2022, 71, 49–56. [Google Scholar] [CrossRef]
- Gandara e Silva, P.P.; Nobre, C.R.; Resaffe, P.; Seabra Pereira, C.D.; Gusmao, F. Leachate from microplastics impairs larval development in brown mussels. Water Res. 2016, 106, 364–370. [Google Scholar] [CrossRef]
- Suzuki, M.; Suzuki, S.; Matsui, M.; Hiraki, Y.; Kawano, F.; Shibayama, K. Genome sequence of a strain of the human pathogenic bacterium Pseudomonas alcaligenes that caused bloodstream infection. Genome Announc. 2013, 1, e00919-13. [Google Scholar] [CrossRef] [Green Version]
- Elkarmi, A.; Abu-Elteen, K.; Khader, M. Modeling the biodegradation efficiency and growth of Pseudomonas alcaligenes utilizing 2,4-dichlorophenol as a carbon source pre- and post-exposure to UV radiation. Jordan J. Biol. Sci. 2008, 1, 7–11. [Google Scholar]
- Bisutti, I.; Hilke, I.; Raessler, M. Determination of total organic carbon—An overview of current methods. TrAC Trends Anal. Chem. 2004, 23, 716–726. [Google Scholar] [CrossRef]
- Tian, X.; Zhao, C.; Ji, X.; Feng, T.; Liu, Y.; Bian, D. The correlation analysis of TOC and CODCr in urban sewage treatment. ICBTE 2019, 136, 06010. [Google Scholar] [CrossRef]
- Baker, A.; Cumberland, S.; Hudson, N. Dissolved and total organic and inorganic carbon in some British rivers. Area 2008, 40, 117–127. [Google Scholar] [CrossRef]
- Keller, B.O.; Sui, J.; Young, A.B.; Whittal, R.M. Interferences and contaminants encountered in modern mass spectrometry. Anal. Chim. Acta 2008, 624, 71–81. [Google Scholar] [CrossRef]
- Wang, S.-C.; Gao, Z.-Y.; Liu, F.-F.; Chen, S.-Q.; Liu, G.-Z. Effects of polystyrene and triphenyl phosphate on growth, photosynthesis and oxidative stress of Chaetoceros meülleri. Sci. Total Environ. 2021, 797, 149180. [Google Scholar] [CrossRef]
- García Ibarra, V.; Sendón, R.; García-Fonte, X.-X.; Paseiro Losada, P.; Rodríguez Bernaldo de Quirós, A. Migration studies of butylated hydroxytoluene, tributyl acetylcitrate and dibutyl phthalate into food simulants. J. Sci. Food Agric. 2019, 99, 1586–1595. [Google Scholar] [CrossRef]
- De-la-Torre, G.E.; Dioses-Salinas, D.C.; Pizarro-Ortega, C.I.; Saldaña-Serrano, M. Global distribution of two polystyrene-derived contaminants in the marine environment: A review. Mar. Pollut. Bull. 2020, 161, 111729. [Google Scholar] [CrossRef]
- Jung, M.R.; Horgen, F.D.; Orski, S.V.; Rodriguez, V.; Beers, K.L.; Balazs, G.H.; Jones, T.T.; Work, T.M.; Brignac, K.C.; Royer, S.-J.; et al. Validation of ATR FT-IR to identify polymers of plastic marine debris, including those ingested by marine organisms. Mar. Pollut. Bull. 2008, 127, 704–716. [Google Scholar] [CrossRef]
- Subramani, M.; Sepperumal, U. FTIR analysis of bacterial mediated chemical changes in polystyrene foam. Ann. Biol. Res. 2016, 7, 55–61. [Google Scholar]
- Bode, H.B.; Zeeck, A.; Jendrossek, D. Physiological and chemical investigations into microbial degradation of synthetic poly(cis-1,4-isoprene). Appl. Environ. Microbiol. 2000, 66, 3680–3685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mooney, A.; Ward, P.G.; O’Connor, K.E. Microbial degradation of styrene: Biochemistry, molecular genetics, and perspectives for biotechnological applications. Appl. Microbiol. Biotechnol. 2006, 72, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Ai, H.; Chen, Y.; Zhang, Z.; Zeng, P.; Kang, L.; Li, W.; Gu, W.; He, Q.; Li, H. Phytoplankton response to polystyrene microplastics: Perspective from an entire growth period. Chemosphere 2018, 208, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Miloloža, M.; Bule, K.; Ukić, Š.; Cvetnić, M.; Bolanča, T.; Kušić, H.; Ocelić Bulatović, V.; Kučić Grgić, D. Ecotoxicological determination of microplastic toxicity on algae Chlorella sp.: Response surface modeling approach. Water Air Soil Pollut. 2021, 232, 327. [Google Scholar] [CrossRef]






| Experiment No. | pH/- | T/°C | MP Size/µm | γMP/mg/L | Agitation Speed/rpm | OD/- | γGLU/mg/L |
|---|---|---|---|---|---|---|---|
| P1-1; P2-1 | 8 | 15 | 600 | 1000 | 100 | 0.5 | 0 |
| P1-2; P2-2 | 6 | 15 | 200 | 50 | 100 | 0.1 | 0 |
| P1-3; P2-3 | 6 | 15 | 200 | 1000 | 200 | 0.5 | 100 |
| P1-4; P2-4 | 8 | 15 | 600 | 50 | 200 | 0.1 | 100 |
| P1-5; P2-5 | 6 | 25 | 600 | 50 | 100 | 0.5 | 100 |
| P1-6; P2-6 | 8 | 25 | 200 | 1000 | 100 | 0.1 | 100 |
| P1-7; P2-7 | 6 | 25 | 600 | 1000 | 200 | 0.1 | 0 |
| P1-8; P2-8 | 8 | 25 | 200 | 50 | 200 | 0.5 | 0 |
| Bacillus cereus | Pseudomonas alcaligenes | ||||||
|---|---|---|---|---|---|---|---|
| Experiment No. | γMP/mg/L | MP Size/µm | Agitation Speed/rpm | Experiment No. | Agitation Speed/rpm | MP Size/µm | OD/- |
| M1-1 | 1000 | 400 | 100 | M2-1 | 200 | 400 | 0.1 |
| M1-2 | 50 | 400 | 200 | M2-2 | 100 | 400 | 0.5 |
| M1-3 | 1000 | 200 | 150 | M2-3 | 200 | 200 | 0.3 |
| M1-4 | 50 | 200 | 100 | M2-4 | 100 | 200 | 0.1 |
| M1-5 | 500 | 400 | 100 | M2-5 | 150 | 400 | 0.1 |
| M1-6 | 50 | 600 | 150 | M2-6 | 100 | 600 | 0.3 |
| M1-7 | 50 | 600 | 100 | M2-7 | 100 | 600 | 0.1 |
| M1-8 | 500 | 200 | 100 | M2-8 | 150 | 200 | 0.1 |
| M1-9 | 500 | 600 | 100 | M2-9 | 150 | 600 | 0.1 |
| M1-10 | 1000 | 200 | 200 | M2-10 | 200 | 200 | 0.5 |
| M1-11 | 1000 | 200 | 100 | M2-11 | 200 | 200 | 0.1 |
| M1-12 | 50 | 200 | 150 | M2-12 | 100 | 200 | 0.3 |
| M1-13 | 500 | 400 | 200 | M2-13 | 150 | 400 | 0.5 |
| M1-14 | 500 | 400 | 150 | M2-14 | 150 | 400 | 0.3 |
| M1-15 | 1000 | 600 | 150 | M2-15 | 200 | 600 | 0.3 |
| M1-16 | 50 | 600 | 200 | M2-16 | 100 | 600 | 0.5 |
| M1-17 | 50 | 200 | 200 | M2-17 | 100 | 200 | 0.5 |
| M1-18 | 1000 | 600 | 200 | M2-18 | 200 | 600 | 0.5 |
| M1-19 | 1000 | 600 | 100 | M2-19 | 200 | 600 | 0.1 |
| M1-20 | 500 | 600 | 150 | M2-20 | 150 | 600 | 0.3 |
| M1-21 | 500 | 600 | 200 | M2-21 | 150 | 600 | 0.5 |
| M1-22 | 1000 | 400 | 200 | M2-22 | 200 | 400 | 0.5 |
| M1-23 | 1000 | 400 | 150 | M2-23 | 200 | 400 | 0.3 |
| M1-24 | 50 | 400 | 100 | M2-24 | 100 | 400 | 0.1 |
| M1-25 | 50 | 400 | 150 | M2-25 | 100 | 400 | 0.3 |
| M1-26 | 500 | 200 | 150 | M2-26 | 150 | 200 | 0.3 |
| M1-27 | 500 | 200 | 200 | M2-27 | 150 | 200 | 0.5 |
| Bacillus cereus | Pseudomonas alcaligenes | |||||
|---|---|---|---|---|---|---|
| Factors | γMP | MP Size | Agitation Speed | Agitation Speed | MP Size | OD |
| Sum of Squares | 0.07 | 0.68 | 0.58 | 1.88 | 0.51 | 0.49 |
| DF * | 1 | 1 | 1 | 1 | 1 | 1 |
| F-value | 128.82 | 1243.26 | 1058.59 | 46.93 | 12.72 | 12.22 |
| p-value | 0.002 | 0.000 | 0.000 | 0.006 | 0.038 | 0.040 |
| Contribution/% | 5.03 | 48.52 | 41.31 | 14.48 | 14.48 | 13.91 |
| Experiment No. | INH/% | EC20/% |
|---|---|---|
| M1-24 | 40.11 | 28.57 |
| M2-23 | 46.79 | 27.40 |
| Bacterium | Applied Model | Statistical Analysis | Influential Model Factors | Influential Parameters | |||||
|---|---|---|---|---|---|---|---|---|---|
| Model | Coefficients | ||||||||
| R2 | R2adj | F | p | Coefficient Value | p | ||||
| Bacillus cereus | MODEL I | 0.8616 | 0.8435 | 47.71 | 0.000 | a0 = 7.55 | 0.000 | agitation speed | |
| a1 = 1.34 × 10−4 | 0.304 | ||||||||
| a2 = −9.02 × 10−5 | 0.107 | ||||||||
| a3 = −6.03 × 10−3 | 0.000 | X3 | |||||||
| MODEL II | 0.9108 | 0.8636 | 19.29 | 0.000 | a0 = 8.16 | 0.000 | agitation speed | ||
| a1 = 1.30 × 10−3 | 0.271 | ||||||||
| a2 = −1.57 × 10−5 | 0.091 | ||||||||
| a3 = −0 × 02 | 0.000 | X3 | |||||||
| a4 = 1.87 × 10−7 | 0.550 | ||||||||
| a5 = −1.02 × 10−6 | 0.732 | ||||||||
| a6 = −8.38 × 10−7 | 0.504 | ||||||||
| a7 = −1.38 × 10−6 | 0.198 | ||||||||
| a8 = −2.23 × 10−8 | 0.905 | ||||||||
| a9 = 4.25 × 10−5 | 0.020 | X32 | |||||||
| Pseudomonas alcaligenes | MODEL I | 0.4287 | 0.3542 | 5.75 | 0.004 | a0 = 7.44 | 0.000 | agitation speed and OD | |
| a1 = 3.87 × 10−3 | 0.011 | X1 | |||||||
| a2 = −3.67 × 10−4 | 0.304 | ||||||||
| a3 = 1.10 | 0.008 | X3 | |||||||
| MODEL II | 0.9076 | 0.8587 | 18.55 | 0.000 | a0 = 3.49 | 0.000 | agitation speed, MP size and OD | ||
| a1 = 4.98 × 10−2 | 0.000 | X1 | |||||||
| a2 = 4.37 × 10−4 | 0.038 | X2 | |||||||
| a3 = 6.93 | 0.000 | X3 | |||||||
| a4 = 2.25 × 10−6 | 0.581 | ||||||||
| a5 = −1.04 × 10−2 | 0.018 | X1· X3 | |||||||
| a6 = 1.60 × 10−3 | 0.127 | ||||||||
| a7 = −1.46 × 10−4 | 0.000 | X12 | |||||||
| a8 = −2.03 × 10−6 | 0.169 | ||||||||
| a9 = −8.32 | 0.000 | X32 | |||||||
| Bacillus cereus | Pseudomonas alcaligenes | |||||
|---|---|---|---|---|---|---|
| Factor | γMP/mg/L | MP Size/µm | Agitation Speed/rpm | Agitation Speed/rpm | MP Size/µm | OD/- |
| Value | 66.20 | 413.29 | 100.45 | 161.08 | 334.73 | 0.35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miloloža, M.; Ukić, Š.; Cvetnić, M.; Bolanča, T.; Kučić Grgić, D. Optimization of Polystyrene Biodegradation by Bacillus cereus and Pseudomonas alcaligenes Using Full Factorial Design. Polymers 2022, 14, 4299. https://doi.org/10.3390/polym14204299
Miloloža M, Ukić Š, Cvetnić M, Bolanča T, Kučić Grgić D. Optimization of Polystyrene Biodegradation by Bacillus cereus and Pseudomonas alcaligenes Using Full Factorial Design. Polymers. 2022; 14(20):4299. https://doi.org/10.3390/polym14204299
Chicago/Turabian StyleMiloloža, Martina, Šime Ukić, Matija Cvetnić, Tomislav Bolanča, and Dajana Kučić Grgić. 2022. "Optimization of Polystyrene Biodegradation by Bacillus cereus and Pseudomonas alcaligenes Using Full Factorial Design" Polymers 14, no. 20: 4299. https://doi.org/10.3390/polym14204299
APA StyleMiloloža, M., Ukić, Š., Cvetnić, M., Bolanča, T., & Kučić Grgić, D. (2022). Optimization of Polystyrene Biodegradation by Bacillus cereus and Pseudomonas alcaligenes Using Full Factorial Design. Polymers, 14(20), 4299. https://doi.org/10.3390/polym14204299