CAGW Peptide Modified Biodegradable Cationic Copolymer for Effective Gene Delivery
Abstract
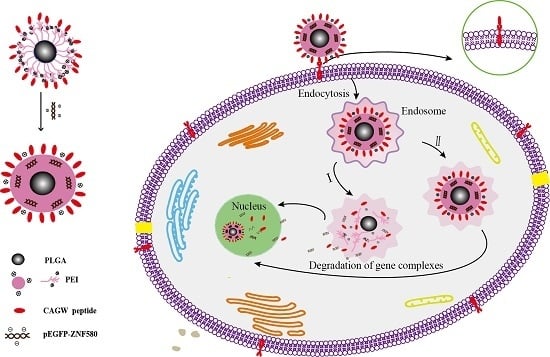
:1. Introduction
2. Material and Methods
2.1. Materials
2.2. Synthesis of Cationic Copolymer PLGA-g-PEI
2.3. Characterization of Cationic Copolymer PLGA-g-PEI
2.4. Preparation and Characterization of Micelles
2.4.1. Preparation of PLGA-g-PEI-g-CAGW Copolymer
2.4.2. Preparation of PLGA-g-PEI-g-CAGW Copolymer Micelles
2.4.3. Characterization of PLGA-g-PEI-g-CAGW Copolymer Micelles
2.5. Preparation of PLGA-g-PEI-g-CAGW/pDNA and PLGA-g-PEI/pDNA Gene Complexes
2.6. Size and Zeta Potential of PLGA-g-PEI-g-CAGW/pDNA and PLGA-g-PEI/pDNA Gene Complexes
2.7. Agarose Gel Electrophoresis
2.8. In Vitro Cytotoxicity of Copolymer Micelles and Gene Complexes
2.9. In Vitro Transfection
2.10. Western Blot Assay
2.11. Wound Healing Assay
2.12. Fluorescence-Activated Cell Sorting (FACS) Analysis
2.13. Statistical Analyses
3. Results
3.1. Synthesis of PLGA-g-PEI and PLGA-g-PEI-g-CAGW Copolymers
3.2. Size and Zeta Potential of PLGA-g-PEI/pDNA and PLGA-g-PEI-g-CAGW/pDNA Gene Complexes
3.3. Agarose Gel Electrophoresis
3.4. In Vitro Cytotoxicity
3.5. In Vitro Transfection
3.6. Western Blot Analysis
3.7. Wound Healing Assay
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ren, X.; Feng, Y.; Guo, J.; Wang, H.; Li, Q.; Yang, J.; Hao, X.; Lv, J.; Ma, N.; Li, W. Surface modification and endothelialization of biomaterials as potential scaffolds for vascular tissue engineering applications. Chem. Soc. Rev. 2015, 44, 5680–5742. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Yao, F.; Li, Q.; Khan, M.; Ren, X.; Feng, Y.; Huang, J.; Zhang, W. Regulation of the endothelialization by human vascular endothelial cells by ZNF580 gene complexed with biodegradable microparticles. Biomaterials 2014, 35, 7133–7145. [Google Scholar] [CrossRef] [PubMed]
- Uttayarat, P.; Perets, A.; Li, M.; Pimtom, P.; Stachlek, S.; Alferiev, I.; Composto, R.; Levy, R.; Lelkes, P.I. Micropatterning of three-dimensional electrospun polyurethane vascular grafts. Acta Biomater. 2010, 6, 4229–4237. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W.; Zhang, J.; Sun, W.; Zhang, R.; Gu, H. Fabrication of a novel polymer-free nanostructured drug-eluting coating for cardiovascular stents. ACS Appl. Mater. Interfaces 2013, 5, 10337–10345. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhao, H.; Behl, M.; Lendlein, A.; Guo, J.; Yang, D. Grafting of poly(ethylene glycol) monoacrylates on polycarbonateurethane by UV initiated polymerization for improving hemocompatibility. J. Mater. Sci. Mater. Med. 2013, 24, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Neffe, A.T.; von Ruesten-Lange, M.; Braune, S.; Luetzow, K.; Roch, T.; Richau, K.; Jung, F.; Lendlein, A. Poly(ethylene glycol) grafting to poly(ether imide) membranes: Influence on protein adsorption and thrombocyte adhesion. Macromol. Biosci. 2013, 13, 1720–1729. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, K.; Suda, T.; Zhang, G.; Liu, D. Advances in gene delivery systems. Pharm. Med. 2011, 25, 293–306. [Google Scholar] [CrossRef]
- Zheng, N.; Song, Z.Y.; Liu, Y.; Yin, L.C.; Cheng, J.J. Gene delivery into isolated Arabidopsis thaliana protoplasts and intact leaves using cationic, α-helical polypeptide. Front. Chem. Sci. Eng. 2017. [Google Scholar] [CrossRef]
- Bonamassa, B.; Liu, D. Nonviral gene transfer as a tool for studying transcription regulation of xenobiotic metabolizing enzymes. Adv. Drug Deliv. Rev. 2010, 62, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Yao, F.; Huang, J.; Han, G.; Li, Q.; Khan, M.; Feng, Y.; Zhang, W. Proliferation and migration of human vascular endothelial cells mediated by ZNF580 gene complexed with mPEG-b-P (MMD-co-GA)-g-PEI microparticles. J. Mater. Chem. B 2014, 2, 1825–1837. [Google Scholar] [CrossRef]
- Merdan, T.; Kopec̆ek, J.; Kissel, T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv. Drug Deliv. Rev. 2002, 54, 715–758. [Google Scholar] [CrossRef]
- Lv, J.; Hao, X.; Yang, J.; Feng, Y.; Behl, M.; Lendlein, A. Selfa—Assembly of polyethylenimine—Modified biodegradable complex micelles as gene transfer vector for proliferation of endothelial cells. Macromol. Chem. Phys. 2014, 215, 2463–2472. [Google Scholar] [CrossRef]
- Funhoff, A.M.; Van Nostrum, C.F.; Lok, M.C.; Fretz, M.M.; Crommelin, D.J.; Hennink, W.E. Poly(3-guanidinopropyl methacrylate): A novel cationic polymer for gene delivery. Bioconjug. Chem. 2004, 15, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zeng, F.; Jian, H.; Wu, S. Grafting zwitterionic polymer chains onto PEI as a convenient strategy to enhance gene delivery performance. Polym. Chem. 2013, 4, 5810–5818. [Google Scholar] [CrossRef]
- Calarco, A.; Bosetti, M.; Margarucci, S.; Fusaro, L.; Nicoli, E.; Petillo, O.; Cannas, M.; Galderisi, U.; Peluso, G. The genotoxicity of PEI-based nanoparticles is reduced by acetylation of polyethylenimine amines in human primary cells. Toxicol. Lett. 2013, 218, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Zaric, V.; Weltin, D.; Erbacher, P.; Remy, J.S.; Behr, J.P.; Stephan, D. Effective polyethylenimine-mediated gene transfer into human endothelial cells. J. Gene Med. 2004, 6, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Furgeson, D.Y.; Yockman, J.W.; Janat, M.M.; Kim, S. Tumor efficacy and biodistribution of linear polyethylenimine-cholesterol/DNA complexes. Mol. Ther. 2004, 9, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.S.; Kang, H.S.; Park, J.Y.; Park, T.G.; Han, S.H.; Chang, I.S. New micelle-like polymer aggregates made from PEI-PLGA diblock copolymers: Micellar characteristics and cellular uptake. Biomaterials 2003, 24, 2053–2059. [Google Scholar] [CrossRef]
- Zhao, F.; Yin, H.; Li, J. Supramolecular self-assembly forming a multifunctional synergistic system for targeted co-delivery of gene and drug. Biomaterials 2014, 35, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Wang, X.; Hu, Y.; Hu, H.; Wu, D.C.; Xu, F.J. Bioreducible POSS-cored star-shaped polycation for efficient gene delivery. ACS Appl. Mater. Interfaces 2013, 6, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Hu, Y.; Yin, L.; Tang, C.; Yin, C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials 2010, 31, 3657–3666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.F.; Yi, W.J.; Wang, B.; Zhang, J.; Ren, L.; Chen, Q.M.; Guo, L.D.; Yu, X.Q. Linear polycations by ring-opening polymerization as non-viral gene delivery vectors. Biomaterials 2013, 34, 5391–5401. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Wang, Z.; Song, L.; Zhao, Q.; Zhang, J.; Li, D.; Wang, S.; Han, J.; Zheng, X.; Yang, Z.; et al. Endothelialization and patency of RGD-functionalized vascular grafts in a rabbit carotid artery model. Biomaterials 2012, 33, 2880–2891. [Google Scholar] [CrossRef] [PubMed]
- Hou, B.; Zheng, B.; Yang, W.; Dong, C.; Wang, H.; Chang, J. Construction of near infrared light triggered nanodumbbell for cancer photodynamic therapy. J. Colloid Interface Sci. 2017, 494, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Li, Q.; Lv, J.; Yu, L.; Ren, X.; Zhang, L.; Feng, Y.; Zhang, W. CREDVW-linked polymeric micelles as a targeting gene transfer vector for selective transfection and proliferation of endothelial cells. ACS Appl. Mater. Interfaces 2015, 7, 12128–12140. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Guo, L.; Yu, Y.; Chen, Z.; Zhou, R.; Yuan, Z. Peptide REDV-modified polysaccharide hydrogel with endothelial cell selectivity for the promotion of angiogenesis. J. Biomed. Mater. Res. A 2015, 103, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, Y.; Yang, J.; Guo, J.; Zhang, W. Targeting REDV peptide functionalized polycationic gene carrier for enhancing the transfection and migration capability of human endothelial cells. J. Mater. Chem. B 2015, 3, 3379–3391. [Google Scholar] [CrossRef]
- Li, Q.; Hao, X.; Lv, J.; Ren, X.; Zhang, K.; Ullah, I.; Feng, Y.; Shi, C.; Zhang, W. Mixed micelles obtained by co-assembling comb-like and grafting copolymers as gene carriers for efficient gene delivery and expression in endothelial cells. J. Mater. Chem. B 2017, 5, 1673–1687. [Google Scholar] [CrossRef]
- Wang, H.; Li, Q.; Yang, J.; Guo, J.; Ren, X.; Feng, Y.; Zhang, W. Comb-shaped polymer grafted with REDV peptide, PEG and PEI as targeting gene carrier for selective transfection of human endothelial cells. J. Mater. Chem. B 2017, 5, 1408–1422. [Google Scholar] [CrossRef]
- Lv, J.; Hao, X.; Li, Q.; Akpanyung, M.; Nejjari, A.; Neve, A.L.; Ren, X.; Feng, Y.; Shi, C.; Zhang, W. Star-shaped copolymer grafted PEI and REDV as a gene carrier to improve migration of endothelial cells. Biomater. Sci. 2017, 5, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Yang, J.; Shi, C.; Lv, J.; Feng, Y.; Zhang, W. Surface tailoring for selective endothelialization and platelet inhibition via a combination of SI-ATRP and click chemistry using Cys-Al-Gly-peptide. Acta Biomater. 2015, 20, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hao, X.; Li, Q.; Akpanyung, M.; Nejjari, A.; Neve, A.L.; Ren, X.; Guo, J.; Feng, Y.; Shi, C.; et al. CAGW Peptide-and PEG-modified gene carrier for selective gene delivery and promotion of angiogenesis in HUVECs in vivo. ACS Appl. Mater. Interfaces 2017, 9, 4485–4497. [Google Scholar] [CrossRef] [PubMed]
- LeBaron, R.G.; Athanasiou, K.A. Extracellular matrix cell adhesion peptides: Functional applications in orthopedic materials. Tissue Eng. 2000, 6, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Goessler, U.R.; Bugert, P.; Bieback, K.; Stern-straeter, J.; Bran, G.; Hörmann, K.; Riedel, F. Integrin expression in stem cells from bone marrow and adipose tissue during chondrogenic differentiation. Int. J. Mol. Med. 2008, 21, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Chen, X.; Gellman, S.H.; Masters, K.S. Nylon-3 Polymers that enable selective culture of endothelial cells. J. Am. Chem. Soc. 2013, 135, 16296–16299. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tan, T.T.Y.; Yuan, S.; Choong, C. Multifunctional P(PEGMA)—REDV Conjugated Titanium Surfaces for Improved Endothelial Cell Selectivity and Hemocompatibility. J. Mater. Chem. B 2013, 1, 157–167. [Google Scholar] [CrossRef]
- Shi, C.; Li, Q.; Zhang, W.; Feng, Y.; Ren, X. REDV Peptide Conjugated Nanoparticles/pZNF580 Complexes for Actively Targeting Human Vascular Endothelial Cells. ACS Appl. Mater. Interfaces 2015, 7, 20389–20399. [Google Scholar] [CrossRef] [PubMed]
- Kanie, K.; Narita, Y.; Zhao, Y.; Kuwabara, F.; Satake, M.; Honda, S.; Kaneko, H.; Yoshioka, T.; Okochi, M.; Honda, H.; et al. Collagen Type IV-Specific Tripeptides for Selective Adhesion of Endothelial and Smooth Muscle Cells. Biotechnol. Bioeng. 2012, 109, 1808–1816. [Google Scholar] [CrossRef] [PubMed]
- Melchiorri, A.J.; Hibino, N.; Fisher, J.P. Strategies and techniques to enhance the in situ endothelialization of small-diameter biodegradable polymeric vascular grafts. Tissue Eng. Part B 2013, 19, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Loh, X.J.; Zhang, Z.X.; Mya, K.Y.; Wu, Y.L.; He, C.B.; Li, J. Efficient gene delivery with paclitaxel-loaded DNA-hybrid polyplexes based on cationicpolyhedral oligomeric silsesquioxanes. J. Mater. Chem. 2010, 20, 10634–10642. [Google Scholar] [CrossRef]
- Hu, Y.; Zhu, Y.; Yang, W.T.; Xu, F.J. New star-shaped carriers composed of β-cyclodextrin cores and disulfide-linked poly(glycidyl methacrylate) derivative arms with plentiful flanking secondary amine and hydroxyl groups for highly efficient gene delivery. ACS Appl. Mater. Interfaces 2013, 5, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Loh, X.J.; Wu, Y.L. Cationic star copolymers based on β-cyclodextrins for efficient gene delivery to mouse embryonic stem cell colonies. Chem. Commun. 2015, 51, 10815–10818. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.; Ekholm, A.; Nybom, H.; Renvert, S.; Widen, C.; Rumpunen, K. Effects of plantago major L leaf extracts on oral epithelial cells in a scratch assay. J. Ethnopharmacol. 2012, 141, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, F.; Narita, Y.; Yamawaki-Ogata, A.; Kanie, K.; Kato, R.; Satake, M.; Kaneko, H.; Oshima, H.; Usui, A.; Ueda, Y. Novel small-caliber vascular grafts with trimeric Peptide for acceleration of endothelialization. Thorac. Surg. 2012, 93, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Weljie, A.M.; Vogel, H.J. Tryptophan fluorescence quenching by methionine and selenomethionine residues of calmodulin: Orientation of peptide and protein binding. Biochemistry 1998, 37, 3187–3195. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, P.; Tang, L.; Sun, P.; Liu, W.; Sun, P.; Zuo, A.; Liang, D. Temperature-tuned DNA condensation and gene transfection by PEI-g-(PMEO2MA-b-PHEMA) copolymer-based nonviral vectors. Biomaterials 2010, 31, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Huang, Y.; Wei, T.; Zhang, W.; Wang, W.; Lin, G.; Zhang, X.; Kumar, A.; Du, Q.; Xing, J.; et al. Amphiphilic and biodegradable methoxy polyethylene glycol-block-(polycaprolactone-graft-poly(2-(dimethylamino) ethyl methacrylate)) as an effective gene carrier. Biomaterials 2011, 32, 879–889. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Y.; Cheng, G.; Xie, L.; Nie, Y.; He, B.; Gu, Z.W. Polyethyleneimine/DNA polyplexes with reduction-sensitive hyaluronic acid derivatives shielding for targeted gene delivery. Biomaterials 2013, 34, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.W.; Fitzgerald, M.; Clemons, T.D.; House, M.J.; Padman, B.S.; Shaw, J.A.; Saunders, M.; Harvey, A.R.; Zdyrko, B.; Luzinov, I. Multimodal analysis of PEI-mediated endocytosis of nanoparticles in neural cells. ACS Nano 2011, 5, 8640–8648. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.; Li, Y.; Ahlemeyer, B.; Krieglstein, J.; Kissel, T. In vitro cytotoxicity testing of polycations: Influence of polymer structure on cell viability and hemolysis. Biomaterials 2003, 24, 1121–1131. [Google Scholar] [CrossRef]
- Jorge, A.F.; Dias, R.S.; Pereira, J.C.; Pais, A.A. DNA condensation by pH-responsive polycations. Biomacromolecules 2010, 11, 2399–2406. [Google Scholar] [CrossRef] [PubMed]
- Kunath, K.; Harpe, V.A.; Fischer, D.; Petersen, H.; Ulrich, B.; Voigt, K.; Kissel, T. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: Comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J. Control. Release 2003, 89, 113–125. [Google Scholar] [CrossRef]
- Moon, H.H.; Joo, M.K.; Mok, H.; Lee, M.; Hwang, K.C.; Kim, S.W.; Jeong, J.H.; Choi, D.; Kim, S.H. MSC-based VEGF gene therapy in rat myocardial infarction model using facial amphipathic bile acid-conjugated polyethyleneimine. Biomaterials 2014, 35, 1744–1754. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, Q.; Yang, X.; Feng, Y.; Ren, X.; Shi, C.; Zhang, W. Multitargeting Gene Delivery Systems for Enhancing the Transfection of Endothelial Cells. Macromol. Rapid Commun. 2016, 37, 1926–1931. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Lin, Q.M.; Zhang, L.M.; Liang, Y.Y.; Xue, W. A star-shaped porphyrin-arginine functionalized poly (l-lysine) copolymer for photo-enhanced drug and gene co-delivery. Biomaterials 2014, 35, 4357–4367. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Samsonova, O.; Sproat, B.; Merkel, O.; Kissel, T. Biophysical characterization of hyper-branched polyethylenimine-graft-polycaprolactone-block-mono-methoxyl-poly(ethylene glycol) copolymers (hy-PEI-PCL-mPEG) for siRNA delivery. J. Control. Release 2011, 153, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Endres, T.; Zheng, M.; Kılıç, A.; Tutowska, A.; Beck-Broichsitter, M.; Renz, H.; Merkel, O.M.; Kissel, T. Amphiphilic biodegradable PEG-PCL-PEI triblock copolymers for FRET-capable in vitro and in vivo delivery of siRNA and quantum dots. Mol. Pharm. 2014, 11, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Yuk, S.H.; Oh, K.S.; Cho, S.H.; Lee, B.S.; Kim, S.Y.; Kwak, B.-K.; King, K.; Kwon, I.C. Glycol chitosan/heparin immobilized iron oxide nanoparticles with a tumor-targeting char-acteristic for magnetic resonance imaging. Biomacromolecules 2011, 12, 2335–2343. [Google Scholar] [CrossRef] [PubMed]
- Panyam, J.; Labhasetwar, V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv. Drug Deliv. Rev. 2003, 55, 329–347. [Google Scholar] [CrossRef]














| N/P Molar Ratio | PLGA-g-PEI | PLGA-g-PEI-g-CAGW |
|---|---|---|
| 0 | 0.460 | 0.454 |
| 2 | 0.349 | 0.306 |
| 5 | 0.411 | 0.550 |
| 10 | 0.406 | 0.362 |
| 20 | 0.286 | 0.267 |
| 30 | 0.400 | 0.332 |
| 40 | 0.373 | 0.792 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duo, X.; Wang, J.; Li, Q.; Neve, A.L.; Akpanyung, M.; Nejjari, A.; Ali, Z.S.S.; Feng, Y.; Zhang, W.; Shi, C. CAGW Peptide Modified Biodegradable Cationic Copolymer for Effective Gene Delivery. Polymers 2017, 9, 158. https://doi.org/10.3390/polym9050158
Duo X, Wang J, Li Q, Neve AL, Akpanyung M, Nejjari A, Ali ZSS, Feng Y, Zhang W, Shi C. CAGW Peptide Modified Biodegradable Cationic Copolymer for Effective Gene Delivery. Polymers. 2017; 9(5):158. https://doi.org/10.3390/polym9050158
Chicago/Turabian StyleDuo, Xinghong, Jun Wang, Qian Li, Agnaldo Luis Neve, Mary Akpanyung, Abdelilah Nejjari, Zaidi Syed Saqib Ali, Yakai Feng, Wencheng Zhang, and Changcan Shi. 2017. "CAGW Peptide Modified Biodegradable Cationic Copolymer for Effective Gene Delivery" Polymers 9, no. 5: 158. https://doi.org/10.3390/polym9050158
APA StyleDuo, X., Wang, J., Li, Q., Neve, A. L., Akpanyung, M., Nejjari, A., Ali, Z. S. S., Feng, Y., Zhang, W., & Shi, C. (2017). CAGW Peptide Modified Biodegradable Cationic Copolymer for Effective Gene Delivery. Polymers, 9(5), 158. https://doi.org/10.3390/polym9050158