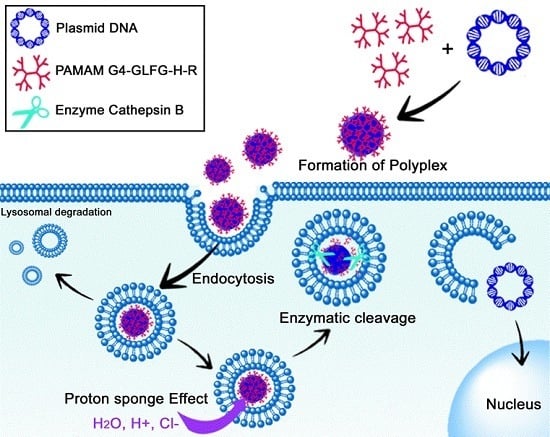
Polyamidoamine (PAMAM) Dendrimers Modified with Cathepsin-B Cleavable Oligopeptides for Enhanced Gene Delivery
Abstract
:1. Introduction
2. Experimental
2.1. Materials
2.2. Synthesis of PAMAM G4-GLFG-H-R
2.3. Gel Retardation Assay and PicoGreen Assay
2.4. Measurement of Dynamic Light Scattering (DLS) and Zeta Potential
2.5. Acid–Base Titration Assay
2.6. Enzymatic Release Test
2.7. Cell Culture and Cytotoxicity Assay of Polymer and Polyplex
2.8. Confocal Microscopy
2.9. Transfection Assay
2.10. Statistical Analysis
3. Result and Discussion
3.1. Synthesis and Characterization
3.2. Gel Retardation Assay and PicoGreen Assay
3.3. Diameter and Zeta Potential of Polyplexes
3.4. Acid–Base Titration
3.5. Plasmid DNA Release Test by Enzyme Cathepsin B
3.6. Cytotoxicity Assay
3.7. Confocal Microscopy
3.8. Transfection Assay
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Miller, K.D.; Siegel, R.L.; Lin, C.C.; Mariotto, A.B.; Kramer, J.L.; Rowland, J.H.; Stein, K.D.; Alteri, R.; Jemal, A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016, 66, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Brannon-Peppas, L.; Blanchette, J.O. Nanoparticle and targeted systems for cancer therapy. Adv. Drug Deliv. Rev. 2004, 56, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Misra, R.; Acharya, S.; Sahoo, S.K. Cancer nanotechnology: Application of nanotechnology in cancer therapy. Drug Discov. Today 2010, 15, 842–850. [Google Scholar] [CrossRef] [PubMed]
- Jayant, R.D.; Sosa, D.; Kaushik, A.; Atluri, V.; Vashist, A.; Tomitaka, A.; Nair, M. Current status of non-viral gene therapy for cns disorders. Expert Opin. Drug Deliv. 2016, 13, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Nayerossadat, N.; Maedeh, T.; Ali, P.A. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012, 1, 27. [Google Scholar] [CrossRef] [PubMed]
- Li, S.D.; Huang, L. Gene therapy progress and prospects: Non-viral gene therapy by systemic delivery. Gene Ther. 2006, 13, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Keeler, A.M.; ElMallah, M.K.; Flotte, T.R. Gene therapy 2017: Progress and future directions. Clin. Transl. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zeng, X.; Liu, M.; Deng, Y.; He, N. Current progress in gene delivery technology based on chemical methods and nano-carriers. Theranostics 2014, 4, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gao, J.; Zhang, C.; Cao, Z.; Cheng, D.; Liu, J.; Shuai, X. Stimuli-responsive polymeric nanocarriers for efficient gene delivery. Top. Curr. Chem. 2017, 375, 27. [Google Scholar] [CrossRef] [PubMed]
- Kaur, D.; Jain, K.; Mehra, N.K.; Kesharwani, P.; Jain, N.K. A review on comparative study of ppi and pamam dendrimers. J Nanopart. Res 2016, 18, 146. [Google Scholar] [CrossRef]
- Esfand, R.; Tomalia, D.A. Poly(amidoamine) (PAMAM) dendrimers: From biomimicry to drug delivery and biomedical applications. Drug Discov. Today 2001, 6, 427–436. [Google Scholar] [CrossRef]
- Kurtoglu, Y.E.; Navath, R.S.; Wang, B.; Kannan, S.; Romero, R.; Kannan, R.M. Poly(amidoamine) dendrimer-drug conjugates with disulfide linkages for intracellular drug delivery. Biomaterials 2009, 30, 2112–2121. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Gupta, U.; Asthana, A.; Jain, N.K. Folate and folate-peg-pamam dendrimers: Synthesis, characterization, and targeted anticancer drug delivery potential in tumor bearing mice. Bioconjug. Chem. 2008, 19, 2239–2252. [Google Scholar] [CrossRef] [PubMed]
- Majoros, I.J.; Myc, A.; Thomas, T.; Mehta, C.B.; Baker, J.R., Jr. Pamam dendrimer-based multifunctional conjugate for cancer therapy: Synthesis, characterization, and functionality. Biomacromolecules 2006, 7, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Boussif, O.; Lezoualch, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo—Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.S.; Bae, Y.M.; Choi, H.; Kong, B.; Choi, I.S.; Choi, J.S. Synthesis of pamam dendrimer derivatives with enhanced buffering capacity and remarkable gene transfection efficiency. Bioconjug. Chem. 2011, 22, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Thuy le, T.; Mallick, S.; Choi, J.S. Polyamidoamine (PAMAM) dendrimers modified with short oligopeptides for early endosomal escape and enhanced gene delivery. Int. J. Pharm. 2015, 492, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jung, J.; Kim, Y.J.; Lee, E.; Choi, J.S. Gene delivery of pamam dendrimer conjugated with the nuclear localization signal peptide originated from fibroblast growth factor 3. Int. J. Pharm. 2014, 459, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Kurtoglu, Y.E.; Mishra, M.K.; Kannan, S.; Kannan, R.M. Drug release characteristics of pamam dendrimer-drug conjugates with different linkers. Int. J. Pharm. 2010, 384, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.J.; Meares, C.F. Cathepsin substrates as cleavable peptide linkers in bioconjugates, selected from a fluorescence quench combinatorial library. Bioconjug. Chem. 1998, 9, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Sloane, B.F.; Yan, S.Q.; Podgorski, I.; Linebaugh, B.E.; Cher, M.L.; Mai, J.X.; Cavallo-Medved, D.; Sameni, M.; Dosescu, J.; Moin, K. Cathepsin B and tumor proteolysis: Contribution of the tumor microenvironment. Semin. Cancer Biol. 2005, 15, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Cho, S.S.; You, J.R.; Lee, Y.; Kang, B.D.; Choi, J.S.; Park, J.W.; Suh, Y.L.; Kim, J.A.; Kim, D.K.; et al. Intraperitoneal gene delivery mediated by a novel cationic liposome in a peritoneal disseminated ovarian cancer model. Gene Ther. 2002, 9, 859–866. [Google Scholar] [PubMed]
- Choi, J.S.; Nam, K.; Park, J.Y.; Kim, J.B.; Lee, J.K.; Park, J.S. Enhanced transfection efficiency of pamam dendrimer by surface modification with l-arginine. J. Control. Release 2004, 99, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, S.; Chen, J.; Orr, B.G.; Holl, M.M.B. Cationic polymer intercalation into the lipid membrane enables intact polyplex DNA escape from endosomes for gene delivery. Mol. Pharm. 2016, 13, 1967–1978. [Google Scholar] [CrossRef] [PubMed]
- De la Rica, R.; Aili, D.; Stevens, M.M. Enzyme-responsive nanoparticles for drug release and diagnostics. Adv. Drug Deliv. Rev. 2012, 64, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Gondi, C.S.; Rao, J.S. Cathepsin b as a cancer target. Expert Opin. Ther. Targets 2013, 17, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Glangchai, L.C.; Caldorera-Moore, M.; Shi, L.; Roy, K. Nanoimprint lithography based fabrication of shape-specific, enzymatically-triggered smart nanoparticles. J. Control. Release 2008, 125, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Lee, M. Apoptosis induced by polyethylenimine/DNA complex in polymer mediated gene delivery. Bull. Korean Chem. Soc. 2007, 28, 95–98. [Google Scholar]
- Jones, C.H.; Chen, C.K.; Ravikrishnan, A.; Rane, S.; Pfeifer, B.A. Overcoming nonviral gene delivery barriers: Perspective and future. Mol. Pharm. 2013, 10, 4082–4098. [Google Scholar] [CrossRef] [PubMed]










| Sample | Zeta Potential | Diameter a | Polydispersity (PDI) a |
|---|---|---|---|
| pDNA | –15.46 ± 5.69 | - | - |
| PAMAM G4 | +37.20 ± 1.95 | 237.0 ± 6.3 | 0.07 |
| G4-GLFG-R | +42.63 ± 0.38 | 199.3 ± 3.9 | 0.17 |
| G4-GLFG-H-R | +46.03 ± 0.81 | 177.2 ± 2.6 | 0.17 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Son, S.J.; Song, S.J.; Ha, T.H.; Choi, J.S. Polyamidoamine (PAMAM) Dendrimers Modified with Cathepsin-B Cleavable Oligopeptides for Enhanced Gene Delivery. Polymers 2017, 9, 224. https://doi.org/10.3390/polym9060224
Lee S, Son SJ, Song SJ, Ha TH, Choi JS. Polyamidoamine (PAMAM) Dendrimers Modified with Cathepsin-B Cleavable Oligopeptides for Enhanced Gene Delivery. Polymers. 2017; 9(6):224. https://doi.org/10.3390/polym9060224
Chicago/Turabian StyleLee, Seulgi, Sang Jae Son, Su Jeong Song, Tai Hwan Ha, and Joon Sig Choi. 2017. "Polyamidoamine (PAMAM) Dendrimers Modified with Cathepsin-B Cleavable Oligopeptides for Enhanced Gene Delivery" Polymers 9, no. 6: 224. https://doi.org/10.3390/polym9060224
APA StyleLee, S., Son, S. J., Song, S. J., Ha, T. H., & Choi, J. S. (2017). Polyamidoamine (PAMAM) Dendrimers Modified with Cathepsin-B Cleavable Oligopeptides for Enhanced Gene Delivery. Polymers, 9(6), 224. https://doi.org/10.3390/polym9060224