Characterization of Cell Scaffolds by Atomic Force Microscopy
Abstract
:1. Introduction
2. Atomic Force Microscopy, a Versatile Tool
2.1. AFM as an Imaging Machine
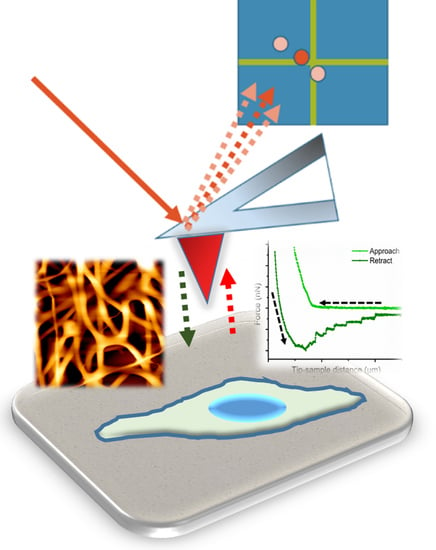
2.2. AFM as a Mechanical Machine
- The approach curve delivers information about the existing repulsive or attractive forces between the tip/colloidal probe and the sample (e.g., electrostatic, van der Waals, hydration, or entropic forces). These type of measurements have been crucial for the understanding of molecular and colloidal interactions [11,36,37,38].
- The second part of the curve, during contact between the cantilever and the sample, provides information about rheology-related properties (e.g., Young´s Modulus, stiffness, relaxation time, and viscosity). The estimation of both the sample stiffness and elastic modulus (E) has been described in [39,40]. In this regard, the Hertz model—in which the contact between two linear, elastic spheres is described—is one of the most commonly used models to calculate the Young’s modulus from an AFM force-distance curve [41]. However, this model presents some limitations, mostly related to the omission of the adhesive forces, which limits its applicability on sticky materials. Alternatively, the Derjaguin-Mfiller-Toporov (DMT) and Johnson-Kendall-Roberts (JKR) theories were developed to overcome such limitations [42]. Also, the employment of indenter geometries different than spheres has given rise to additional adjustments, as with the Sneddon model, which is applied for conical shapes [43] and the derivative equations developed for quadratic pyramids or flat indenters [44,45,46]. In addition, more information can be obtained by keeping close contact between the tip and the material for a certain observation time (tobservation), which is normally denoted as the Dwell time. Depending on the measurement performed, either the Z position of the head (Relaxation) or the load applied (Creep Compliance) are fixed during the contact. This induces the material to undergo structural rearrangement in response to the load-induced deformation which, by extension, allows the obtaining of the compressive moduli and viscosities of the material tested [47].
- Finally, the segment depicting the retraction motion relates to adhesive forces, the existence of tethers, and possible molecular unfolding events. The maximum adhesion (Fadh) parameter, or pull-off force, is indicative of the stickiness of the sample. It is brought by the minimum of the peak in the retraction segment. Additional pulling shows the recovery path followed until achievement of the non-contact state. This tip-sample retraction can take place either via tether formation, in the shape of uniform rupture events distanced by plateaus of zero force variation [48], or when capturing individual molecules/chains, by means of saw-like adhesion peaks to be fitted by a worm-like chain (WLC) model [49,50].
3. AFM and Cell Scaffolds: Literature Review
3.1. Fibres
3.2. Patterned Structures
3.3. Particles
3.4. Hydrogels
3.5. Peptides and RGD Sequence
4. Conclusions and Outlook
- The first part of the plot (approach motion) can be used for determining the presence of repulsive/attractive forces between the tip and the simple (electrostatic, steric, and/or entropic, etc.). Examples of it were already reported in the literature, among others, by Borkovek [24], and Melzak [98]. A similar process is also described by Gentsch et al. in reference [92], and depicted in Figure 3B.
- Attending to the contact segment of the plot (or pause in contact) or studying the deformation of the scaffold under constant force or stress relaxation experiments can describe the rheology of the material for sufficiently large observation (Dwell) times. This was successfully applied to characterize the response of breast carcinoma MCF-7 cells seeded on borosilicate glass substrates [97]. In their work, the authors also presented a new type of AFM imaging called Stress Relaxation imaging, based on the local relaxation times measured after subdividing the cell into multiple domains (mapping in Figure 4B).
- The retraction motion (third segment) yields, in turn, very useful information about adhesive forces and the work required to recover the non-contact state. A good example of it would be the aforementioned “single” force microscopy mode for the stretching of individual coiled chains adhered to the tip (as in the case of proteins) [99,100,101]. Hence, quantification of the occurring intermediate rupture events, as the cantilever moves perpendicularly away from the surface, might deliver a precise fingerprint of the forces which govern the internal arrangement of the coils (Figure 4C).
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Han, D.K.; Park, K.D.; Hubbell, J.A.; Kim, Y.H. Surface characteristics and biocompatibility of lactide-based poly(ethylene glycol) scaffolds for tissue engineering. J. Biomater. Sci. Polym. Ed. 1998, 9, 667–680. [Google Scholar] [PubMed]
- Irvine, D.J.; Ruzette, A.V.G.; Mayes, A.M.; Griffith, L.G. Nanoscale clustering of rgd peptides at surfaces using comb polymers. 2. Surface segregation of comb polymers in polylactide. Biomacromolecules 2001, 2, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Marshall, G.W., Jr.; Balooch, M.; Kinney, J.H.; Marshall, S.J. Atomic force microscopy of conditioning agents on dentin. J. Biomed. Mater. Res. 1995, 29, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Stock, U.A.; Vacanti, J.P. Tissue engineering: Current state and prospects. Annu. Rev. Med. 2001, 52, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Janmey, P.; Wang, Y.-L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Prauzner-Bechcicki, S.; Raczkowska, J.; Madej, E.; Pabijan, J.; Lukes, J.; Sepitka, J.; Rysz, J.; Awsiuk, K.; Bernasik, A.; Budkowski, A.; et al. PDMS substrate stiffness affects the morphology and growth profiles of cancerous prostate and melanoma cells. J. Mech. Behav. Biomed. Mater. 2015, 41, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Vogel, V.; Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Yim, E.K.; Darling, E.M.; Kulangara, K.; Guilak, F.; Leong, K.W. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials 2010, 31, 1299–1306. [Google Scholar] [CrossRef] [PubMed]
- Hinterdorfer, P.; Gruber, H.J.; Kienberger, F.; Kada, G.; Riener, C.; Borken, C.; Schindler, H.J. Surface attachment of ligands and receptors for molecular recognition force microscopy. Colloids Surf. B 2002, 23, 115–123. [Google Scholar] [CrossRef]
- Sugimoto, Y.; Pou, P.; Abe, M.; Jelinek, P.; Perez, R.; Morita, S.; Custance, O. Chemical identification of individual surface atoms by atomic force microscopy. Nature 2007, 446, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Weisenhorn, A.L.; Maivald, P.; Butt, H.J.; Hansma, P.K. Measuring adhesion, attraction, and repulsion between surfaces in liquids with an atomic-force microscope. Phys. Rev. B 1992, 45, 11226–11232. [Google Scholar] [CrossRef]
- Arnoldi, M.; Fritz, M.; Bauerlein, E.; Radmacher, M.; Sackmann, E.; Boulbitch, A. Bacterial turgor pressure can be measured by atomic force microscopy. Phys. Rev. E 2000, 62, 1034–1044. [Google Scholar] [CrossRef]
- Charras, G.T.; Lehenkari, P.P.; Horton, M.A. Atomic force microscopy can be used to mechanically stimulate osteoblasts and evaluate cellular strain distributions. Ultramicroscopy 2001, 86, 85–95. [Google Scholar] [CrossRef]
- Fotiadis, D.; Scheuring, S.; Müller, S.A.; Engel, A.; Müller, D.J. Imaging and manipulation of biological structures with afm. Micron 2002, 33, 385–397. [Google Scholar] [CrossRef]
- Leporatti, S.; Gerth, A.; Kohler, G.; Kohlstrunk, B.; Hauschildt, S.; Donath, E. Elasticity and adhesion of resting and lipopolysaccharide-stimulated macrophages. FEBS Lett. 2006, 580, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.-J.; Cappella, B.; Kappl, M. Force measurements with the atomic force microscope: Technique, interpretation and applications. Surf. Sci. Rep. 2005, 59, 1–152. [Google Scholar] [CrossRef]
- Leckband, D. Novel recognition mechanisms in biological adhesion. Curr. Opin. Colloid Interface Sci. 2001, 6, 498–505. [Google Scholar] [CrossRef]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic force microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed]
- Alessandrini, A.; Facci, P. AFM: A versatile tool in biophysics. Meas. Sci. Technol. 2005, 16, R65–R92. [Google Scholar] [CrossRef]
- Moreno-Flores, S.; Toca-Herrera, J.L. Hybridizing Surface Probe Microscopes: Toward a Full Description of the Meso- and Nanoworlds; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- García, R.; Tamayo, J.; San Paulo, A. Phase contrast and surface energy hysteresis in tapping mode scanning force microscopy. Surf. Interface Anal. 1999, 27, 312–316. [Google Scholar] [CrossRef]
- Radmacher, M.; Tillmann, R.W.; Fritz, M.; Gaub, H.E. From molecules to cells: Imaging soft samples with the atomic force microscope. Science 1992, 257, 1900–1905. [Google Scholar] [CrossRef] [PubMed]
- Martin-Jimenez, D.; Chacon, E.; Tarazona, P.; Garcia, R. Atomically resolved three-dimensional structures of electrolyte aqueous solutions near a solid surface. Nat. Commun. 2016, 7, 12164. [Google Scholar] [CrossRef] [PubMed]
- Borkovec, M.; Szilagyi, I.; Popa, I.; Finessi, M.; Sinha, P.; Maroni, P.; Papastavrou, G. Investigating forces between charged particles in the presence of oppositely charged polyelectrolytes with the multi-particle colloidal probe technique. Adv. Colloid Interface Sci. 2012, 179–182, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Puech, P.H.; Poole, K.; Knebel, D.; Muller, D.J. A new technical approach to quantify cell-cell adhesion forces by AFM. Ultramicroscopy 2006, 106, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Bornschlögl, T.; Rief, M. Single-molecule protein unfolding and refolding using atomic force microscopy. In Methods in Molecular Biology; Springer: Berlin, Germany, 2011; Volume 783, pp. 233–250. [Google Scholar]
- Fisher, T.E.; Oberhauser, A.F.; Carrion-Vazquez, M.; Marszalek, P.E.; Fernandez, J.M. The study of protein mechanics with the atomic force microscope. Trends Biochem. Sci. 1999, 24, 379–384. [Google Scholar] [CrossRef]
- Florin, E.L.; Moy, V.T.; Gaub, H.E. Adhesion forces between individual ligand-receptor pairs. Science 1994, 264, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Hinterdorfer, P.; Dufrêne, Y.F. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 2006, 3, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Melzak, K.A.; Lázaro, G.R.; Hernández-Machado, A.; Pagonabarraga, I.; Cárdenas Díaz De Espada, J.M.; Toca-Herrera, J.L. AFM measurements and lipid rearrangements: Evidence from red blood cell shape changes. Soft Matter 2012, 8, 7716–7726. [Google Scholar] [CrossRef]
- Radmacher, M. Measuring the elastic properties of living cells by the atomic force microscope. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2002; Volume 2002, pp. 67–90. [Google Scholar]
- Scheuring, S.; Dufrêne, Y.F. Atomic force microscopy: Probing the spatial organization, interactions and elasticity of microbial cell envelopes at molecular resolution. Mol. Microbiol. 2010, 75, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Pinto, R.; Gong, H.; Vahabikashi, A.; Johnson, M. The effect of the endothelial cell cortex on atomic force microscopy measurements. Biophys. J. 2013, 105, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Ducker, W.A.; Senden, T.J.; Pashley, R.M. Direct measurement of colloidal forces using an atomic force microscope. Nature 1991, 353, 239–241. [Google Scholar] [CrossRef]
- Cumpson, P.J.; Clifford, C.A.; Portoles, J.F.; Johnstone, J.E.; Munz, M. Cantilever spring-constant calibration in atomic force microscopy. Nano Sci. Technol. 2008, 8, 289–314. [Google Scholar]
- JPK. A Practical Guide to AFM Force Spectroscopy and Data Analysis. In JPK Instruments Technical Notes; JPK Instruments AG: Berlin, Germany; Available online: http://www.jpk.com/afm.230.en.html (accessed on 15 May 2017).
- Butt, H.J. Measuring electrostatic, van der waals, and hydration forces in electrolyte solutions with an atomic force microscope. Biophys. J. 1991, 60, 1438–1444. [Google Scholar] [CrossRef]
- García, R.; San Paulo, A. Attractive and repulsive tip-sample interaction regimes in tapping-mode atomic force microscopy. Phys. Rev. B 1999, 60, 4961–4967. [Google Scholar] [CrossRef]
- Dimitriadis, E.K.; Horkay, F.; Maresca, J.; Kachar, B.; Chadwick, R.S. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 2002, 82, 2798–2810. [Google Scholar] [CrossRef]
- Domke, J.; Radmacher, M. Measuring the elastic properties of thin polymer films with the atomic force microscope. Langmuir 1998, 14, 3320–3325. [Google Scholar] [CrossRef]
- Hertz, H. Uber die berührung fester, elastischer körper. J. Reine Angew. Math. 1882, 92, 156–171. [Google Scholar]
- Cappella, B.; Dietler, G. Force-distance curves by atomic force microscopy. Surf. Sci. Rep. 1999, 34, 1–104. [Google Scholar] [CrossRef]
- Sneddon, I.A. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 1965, 3, 47–57. [Google Scholar] [CrossRef]
- JPK. Determining the Elastic Modulus of Biological Samples Using Atomic Force Microscopy. In JPK Instruments Technical Notes; JPK Instruments AG: Berlin, Germany; Available online: http://www.jpk.com/afm.230.en.html (accessed on 15 May 2017).
- Alcaraz, J.; Buscemi, L.; Grabulosa, M.; Trepat, X.; Fabry, B.; Farre, R.; Navajas, D. Microrheology of human lung epithelial cells measured by atomic force microscopy. Biophys. J. 2003, 84, 2071–2079. [Google Scholar] [CrossRef] [Green Version]
- Gimenez, A.; Uriarte, J.J.; Vieyra, J.; Navajas, D.; Alcaraz, J. Elastic properties of hydrogels and decellularized tissue sections used in mechanobiology studies probed by atomic force microscopy. Microsc. Res. Tech. 2017, 80, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Flores, S.; Benitez, R.; Vivanco, M.D.M.; Toca-Herrera, J.L. Stress relaxation and creep on living cells with the atomic force microscope: A means to calculate elastic moduli and viscosities of cell components. Nanotechnology 2010, 21, 44. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Graham, J.S.; Hegedus, B.; Marga, F.; Zhang, Y.; Forgacs, G.; Grandbois, M. Multiple membrane tethers probed by atomic force microscopy. Biophys. J. 2005, 89, 4320–4329. [Google Scholar] [CrossRef] [PubMed]
- Hugel, T.; Seitz, M. The study of molecular interactions by afm force spectroscopy. Macromol. Rapid Commun. 2001, 22, 989–1016. [Google Scholar] [CrossRef]
- Yamamoto, S.; Tsujii, Y.; Fukuda, T. Atomic force microscopic study of stretching a single polymer chain in a polymer brush. Macromolecules 2000, 33, 5995–5998. [Google Scholar] [CrossRef]
- Benítez, R.; Moreno-flores, S.; Bolós, V.J.; Toca-Herrera, J.L. A new automatic contact point detection algorithm for afm force curves. Microsc. Res. Tech. 2013, 76, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Al Rez, M.F.; Binobaid, A.; Alghosen, A.; Mirza, E.H.; Alam, J.; Fouad, H.; Hashem, M.; Alsalman, H.; Almalak, H.M.; Mahmood, A.; et al. Tubular poly(ε-caprolactone)/chitosan nanofibrous scaffold prepared by electrospinning for vascular tissue engineering applications. J. Biomater. Tissue Eng. 2017, 7, 427–436. [Google Scholar] [CrossRef]
- Chen, J.; Dong, R.; Ge, J.; Guo, B.; Ma, P.X. Biocompatible, biodegradable, and electroactive polyurethane-urea elastomers with tunable hydrophilicity for skeletal muscle tissue engineering. ACS Appl. Mater. Interfaces 2015, 7, 28273–28285. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Huang, C.; Ke, Q.; He, C.; Wang, H.; Mo, X. Preparation and characterization of coaxial electrospun thermoplastic polyurethane/collagen compound nanofibers for tissue engineering applications. Colloids Surf. B Biointerfaces 2010, 79, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Stylianou, A.; Yova, D. Surface nanoscale imaging of collagen thin films by atomic force microscopy. Mater. Sci. Eng. C 2013, 33, 2947–2957. [Google Scholar] [CrossRef] [PubMed]
- Farokhi, M.; Mottaghitalab, F.; Hadjati, J.; Omidvar, R.; Majidi, M.; Amanzadeh, A.; Azami, M.; Tavangar, S.M.; Shokrgozar, M.A.; Ai, J. Structural and functional changes of silk fibroin scaffold due to hydrolytic degradation. J. Appl. Polym. Sci. 2014, 131. [Google Scholar] [CrossRef]
- Pillai, M.M.; Gopinathan, J.; Indumathi, B.; Manjoosha, Y.R.; Santosh Sahanand, K.; Dinakar Rai, B.K.; Selvakumar, R.; Bhattacharyya, A. Silk–PVA hybrid nanofibrous scaffolds for enhanced primary human meniscal cell proliferation. J. Membr. Biol. 2016, 249, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Mo, X.; Huang, C.; He, C.; Wang, H. Electrospun scaffolds from silk fibroin and their cellular compatibility. J. Biomed. Mater. Res. A 2010, 93, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Li, Y.; Yang, B.; Xiao, D.; Zhang, S.; Rajulu, A.V.; Kondo, T.; Zhang, L.; Zhou, J. Effect of microcrystal cellulose and cellulose whisker on biocompatibility of cellulose-based electrospun scaffolds. Cellulose 2013, 20, 1911–1923. [Google Scholar] [CrossRef]
- Babolmorad, G.; Emtiazi, G.; Ghaedi, K.; Jodeiri, M. Enhanced PC12 cells proliferation with self-assembled s-layer proteins scaffolds. Appl. Biochem. Biotechnol. 2015, 175, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.; Natalello, A.; Sanii, B.; Vasita, R.; Saracino, G.; Zuckermann, R.N.; Doglia, S.M.; Gelain, F. Synthesis and characterization of designed bmhp1-derived self-assembling peptides for tissue engineering applications. Nanoscale 2013, 5, 704–718. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zheng, H.; Liang, S.; Gao, C. Aligned plla nanofibrous scaffolds coated with graphene oxide for promoting neural cell growth. Acta Biomater. 2016, 37, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Luo, J.; Fang, C.; Xiong, J. Investigation of polylactide/poly(ε-caprolactone)/multi-walled carbon nanotubes electrospun nanofibers with surface texture. RSC Adv. 2015, 5, 99179–99187. [Google Scholar] [CrossRef]
- Chen, C.; Lv, G.; Pan, C.; Song, M.; Wu, C.; Guo, D.; Wang, X.; Chen, B.; Gu, Z. Poly(lactic acid) (PLA) based nanocomposites—a novel way of drug-releasing. Biomed. Mater. 2007, 2, L1–L4. [Google Scholar] [CrossRef] [PubMed]
- Hung, H.S.; Chang, C.H.; Chang, C.J.; Tang, C.M.; Kao, W.C.; Lin, S.Z.; Hsieh, H.H.; Chu, M.Y.; Sun, W.S.; Hsu, S.H. In vitro study of a novel nanogold-collagen composite to enhance the mesenchymal stem cell behavior for vascular regeneration. PLoS ONE 2014, 9, 104019. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cui, T.; Mills, D.K.; Lvov, Y.M.; McShane, M.J. Comparison of selective attachment and growth of smooth muscle cells on gelatin- and fibronectin-coated micropatterns. J. Nanosci. Nanotechnol. 2005, 5, 1809–1815. [Google Scholar] [CrossRef] [PubMed]
- Marszalek, J.E.; Simon, C.G., Jr.; Thodeti, C.; Adapala, R.K.; Murthy, A.; Karim, A. 2.5d constructs for characterizing phase separated polymer blend surface morphology in tissue engineering scaffolds. J. Biomed. Mater. Res. A 2013, 101 A, 1502–1510. [Google Scholar] [CrossRef] [PubMed]
- Dunne, L.W.; Iyyanki, T.; Hubenak, J.; Mathur, A.B. Characterization of dielectrophoresis-aligned nanofibrous silk fibroin-chitosan scaffold and its interactions with endothelial cells for tissue engineering applications. Acta Biomater. 2014, 10, 3630–3640. [Google Scholar] [CrossRef] [PubMed]
- Horimizu, M.; Kawase, T.; Tanaka, T.; Okuda, K.; Nagata, M.; Burns, D.M.; Yoshie, H. Biomechanical evaluation by AFM of cultured human cell-multilayered periosteal sheets. Micron 2013, 48, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, D.; Liu, S.; Fu, Y.; Kou, X.; Wang, X.; Sha, Y.; Gan, Y.; Zhou, Y. Effect of nanostructure of mineralized collagen scaffolds on their physical properties and osteogenic potential. J. Biomed. Nanotechnol. 2014, 10, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Guarino, V.; Veronesi, F.; Marrese, M.; Giavaresi, G.; Ronca, A.; Sandri, M.; Tampieri, A.; Fini, M.; Ambrosio, L. Needle-like ion-doped hydroxyapatite crystals influence osteogenic properties of PCL composite scaffolds. Biomed. Mater. 2016, 11, 015018. [Google Scholar] [CrossRef] [PubMed]
- Firkowska, I.; Godehardt, E.; Giersig, M. Interaction between human osteoblast cells and inorganic two-dimensional scaffolds based on multiwalled carbon nanotubes: A quantitative AFM study. Adv. Funct. Mater. 2008, 18, 3765–3771. [Google Scholar] [CrossRef]
- Baker, S.R.; Banerjee, S.; Bonin, K.; Guthold, M. Determining the mechanical properties of electrospun poly-ε-caprolactone (PCL) nanofibers using AFM and a novel fiber anchoring technique. Mater. Sci. Eng. C 2016, 59, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.; Sigley, J.; Helms, C.C.; Stitzel, J.; Berry, J.; Bonin, K.; Guthold, M. The mechanical properties of dry, electrospun fibrinogen fibers. Mater. Sci. Eng. C 2012, 32, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Spurlin, T.A.; Bhadriraju, K.; Chung, K.H.; Tona, A.; Plant, A.L. The treatment of collagen fibrils by tissue transglutaminase to promote vascular smooth muscle cell contractile signaling. Biomaterials 2009, 30, 5486–5496. [Google Scholar] [CrossRef] [PubMed]
- Podestà, A.; Ranucci, E.; Macchi, L.; Bongiorno, G.; Ferruti, P.; Milani, P. Micro- and nanoscale modification of poly(2-hydroxyethyl methacrylate) hydrogels by afm lithography and nanoparticle incorporation. J. Nanosci. Nanotechnol. 2005, 5, 425–430. [Google Scholar] [CrossRef] [PubMed]
- D'Acunto, M.; Napolitano, S.; Pingue, P.; Giusti, P.; Rolla, P. Fast formation of ripples induced by AFM. A new method for patterning polymers on nanoscale. Mater. Lett. 2007, 61, 3305–3309. [Google Scholar] [CrossRef]
- McCracken, J.M.; Badea, A.; Kandel, M.E.; Gladman, A.S.; Wetzel, D.J.; Popescu, G.; Lewis, J.A.; Nuzzo, R.G. Programming mechanical and physicochemical properties of 3d hydrogel cellular microcultures via direct ink writing. Adv. Healthc. Mater. 2016, 5, 1025–1039. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Bastatas, L.; Matthews, J.; Lee, Y.J. Mechanical responses of cancer cells on nanoscaffolds for adhesion size control. Macromol. Biosci. 2015, 15, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Kim, H.; Lee, J.; Jeon, H.; Ryu, W. Direct modulus measurement of single composite nanofibers of silk fibroin/hydroxyapatite nanoparticles. Compos. Sci. Technol. 2016, 122, 113–121. [Google Scholar] [CrossRef]
- Hong, Z.; Luz, G.M.; Hampel, P.J.; Jin, M.; Liu, A.; Chen, X.; Mano, J.F. Mono-dispersed bioactive glass nanospheres: Preparation and effects on biomechanics of mammalian cells. J. Biomed. Mater. Res. A 2010, 95, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Deligkaris, K.; Tadele, T.S.; Olthuis, W.; van den Berg, A. Hydrogel-based devices for biomedical applications. Sens. Actuators B Chem. 2010, 147, 765–774. [Google Scholar] [CrossRef]
- Abuelfilat, A.Y.; Kim, Y.; Miller, P.; Hoo, S.P.; Li, J.; Chan, P.; Fu, J. Bridging structure and mechanics of three-dimensional porous hydrogel with X-ray ultramicroscopy and atomic force microscopy. RSC Adv. 2015, 5, 63909–63916. [Google Scholar] [CrossRef]
- Credi, C.; Biella, S.; De Marco, C.; Levi, M.; Suriano, R.; Turri, S. Fine tuning and measurement of mechanical properties of crosslinked hyaluronic acid hydrogels as biomimetic scaffold coating in regenerative medicine. J. Mech. Behav. Biomed. Mater. 2014, 29, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Ohya, S.; Kidoaki, S.; Matsuda, T. Poly(n-isopropylacrylamide) (PNIPAM)-grafted gelatin hydrogel surfaces: Interrelationship between microscopic structure and mechanical property of surface regions and cell adhesiveness. Biomaterials 2005, 26, 3105–3111. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zheng, Q.; Wu, Y.; Liu, Y.; Guo, X.; Wu, W. Culture of nucleus pulposus cells from intervertebral disc on self-assembling kld-12 peptide hydrogel scaffold. Mater. Sci. Eng. C 2010, 30, 975–980. [Google Scholar] [CrossRef]
- Cinar, G.; Ceylan, H.; Urel, M.; Erkal, T.S.; Deniz Tekin, E.; Tekinay, A.B.; Dâna, A.; Guler, M.O. Amyloid inspired self-assembled peptide nanofibers. Biomacromolecules 2012, 13, 3377–3387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Secchi, V.; Franchi, S.; Fioramonti, M.; Polzonetti, G.; Iucci, G.; Bochicchio, B.; Battocchio, C. Nanofibers of human tropoelastin-inspired peptides: Structural characterization and biological properties. Mater. Sci. Eng. C 2017, 77, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Bellis, S.L. Advantages of rgd peptides for directing cell association with biomaterials. Biomaterials 2011, 32, 4205–4210. [Google Scholar] [CrossRef] [PubMed]
- Hersel, U.; Dahmen, C.; Kessler, H. RGD modified polymers: Biomaterials for stimulated cell adhesion and beyond. Biomaterials 2003, 24, 4385–4415. [Google Scholar] [CrossRef]
- Sabater i Serra, R.; León-Boigues, L.; Sánchez-Laosa, A.; Gómez-Estrada, L.; Gómez Ribelles, J.L.; Salmeron-Sanchez, M.; Gallego Ferrer, G. Role of chemical crosslinking in material-driven assembly of fibronectin (nano)networks: 2d surfaces and 3d scaffolds. Colloids Surf. B Biointerfaces 2016, 148, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Gentsch, R.; Pippig, F.; Schmidt, S.; Cernoch, P.; Polleux, J.; Börner, H.G. Single-step electrospinning to bioactive polymer nanofibers. Macromolecules 2011, 44, 453–461. [Google Scholar] [CrossRef]
- Moreno-Cencerrado, A.; Iturri, J.; Pecorari, I.; Vivanco, M.D.M.; Sbaizero, O.; Toca-Herrera, J.L. Investigating cell-substrate and cell-cell interactions by means of single-cell-probe force spectroscopy. Microsc. Res. Tech. 2017, 80, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Taubenberger, A.V.; Hutmacher, D.W.; Muller, D.J. Single-cell force spectroscopy, an emerging tool to quantify cell adhesion to biomaterials. Tissue Eng. B Rev. 2014, 20, 40–55. [Google Scholar] [CrossRef] [PubMed]
- McKendry, R.; Theoclitou, M.-E.; Rayment, T.; Abell, C. Chiral discrimination by chemical forcemicroscopy. Nature 1997, 391, 566–568. [Google Scholar]
- Senapati, S.; Lindsay, S. Recent progress in molecular recognition imaging using atomic force microscopy. Acc. Chem. Res. 2016, 49, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Flores, S.; Benitez, R.; Vivanco, M.D.; Toca-Herrera, J.L. Stress relaxation microscopy: Imaging local stress in cells. J. Biomech. 2010, 43, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Melzak, K.A.; Mateescu, A.; Toca-Herrera, J.L.; Jonas, U. Simultaneous measurement of mechanical and surface properties in thermoresponsive, anchored hydrogel films. Langmuir 2012, 28, 12871–12878. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Brockwell, D.J.; Toca-Herrera, J.L.; Blake, A.W.; Smith, D.A.; Radford, S.E.; Clarke, J. Force mode atomic force microscopy as a tool for protein folding studies. Anal. Chim. Acta 2003, 479, 87–105. [Google Scholar] [CrossRef]
- Oberhauser, A.F.; Marszalek, P.E.; Carrion-Vazquez, M.; Fernandez, J.M. Single protein misfolding events captured by atomic force microscopy. Nat. Struct. Biol. 1999, 6, 1025–1028. [Google Scholar] [PubMed]
- Schlierf, M.; Li, H.; Fernandez, J.M. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl. Acad. Sci. USA 2004, 101, 7299–7304. [Google Scholar] [CrossRef] [PubMed]
- Cartagena-Rivera, A.X.; Wang, W.H.; Geahlen, R.L.; Raman, A. Fast, multi-frequency, and quantitative nanomechanical mapping of live cells using the atomic force microscope. Sci. Rep. 2015, 5, 11692. [Google Scholar] [CrossRef] [PubMed]
- Heu, C.; Berquand, A.; Elie-Caille, C.; Nicod, L. Glyphosate-induced stiffening of hacat keratinocytes, a peak force tapping study on living cells. J. Struct. Biol. 2012, 178, 1–7. [Google Scholar] [CrossRef] [PubMed]




© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iturri, J.; Toca-Herrera, J.L. Characterization of Cell Scaffolds by Atomic Force Microscopy. Polymers 2017, 9, 383. https://doi.org/10.3390/polym9080383
Iturri J, Toca-Herrera JL. Characterization of Cell Scaffolds by Atomic Force Microscopy. Polymers. 2017; 9(8):383. https://doi.org/10.3390/polym9080383
Chicago/Turabian StyleIturri, Jagoba, and José L. Toca-Herrera. 2017. "Characterization of Cell Scaffolds by Atomic Force Microscopy" Polymers 9, no. 8: 383. https://doi.org/10.3390/polym9080383
APA StyleIturri, J., & Toca-Herrera, J. L. (2017). Characterization of Cell Scaffolds by Atomic Force Microscopy. Polymers, 9(8), 383. https://doi.org/10.3390/polym9080383