Studying Gene Expression in Irradiated Barley Cultivars: PM19L-like and CML31-like Expression as Possible Determinants of Radiation Hormesis Effect
Abstract
:1. Introduction
2. Materials and Methods
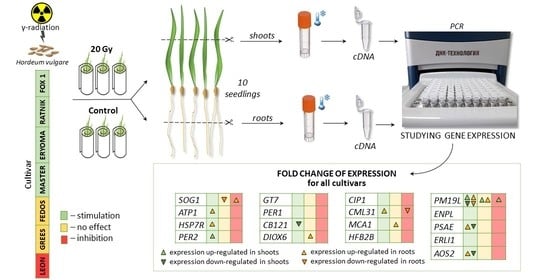
2.1. Barley Cultivars, Seed Irradiation, and Growth Conditions
2.2. Morphological Assessment of Seedlings
2.3. Cultivars Ranking
2.4. Irradiation and Sampling Conditions for Gene Expression Analysis
2.5. qPCR-RT Analysis
2.6. Data Analysis
3. Results
3.1. Ranking of Cultivars According to Radiosensitivity
3.2. Gene Expression Analysis in Roots and Shoots of Irradiated Plants
3.2.1. General Comparison of Gene Expression among Different Organs of Irradiated Cultivars
3.2.2. Differentially Expressed Genes in “No Effect” and γ-Inhibited Cultivars
3.2.3. Differentially Expressed Genes in γ-Stimulated Cultivars
3.2.4. Genes Expressed only for Irradiated or Non-Irradiated Plants
3.2.5. Expression of Putative Target Genes PM19L and CML31
3.3. Correlations between Morphological Traits and Gene Expression
4. Discussion
4.1. Morphological Responses of Seedlings to Seed γ-Irradiation
4.2. Gene Expression in γ-Inhibited and “No Effect” Cultivars
4.3. Gene Expression in γ-Stimulated Cultivars
4.4. Genes Expressed Only for Irradiated or Non-Irradiated Plants
4.5. PM19L and CML31—Putative Target Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Agathokleous, E.; Kitao, M.; Calabrese, E.J. Hormesis: Highly generalizable and beyond laboratory. Trends Plant Sci. 2020. [Google Scholar] [CrossRef]
- Vázquez-Hernández, M.C.; Parola-Contreras, I.; Montoya-Gómez, L.M.; Torres-Pacheco, I.; Schwarz, D.; Guevara-González, R.G. Eustressors: Chemical and physical stress factors used to enhance vegetables production. Sci. Hortic. 2019, 250, 223–229. [Google Scholar] [CrossRef]
- Araújo, S.d.S.; Paparella, S.; Dondi, D.; Bentivoglio, A.; Carbonera, D.; Balestrazzi, A. Physical methods for seed invigoration: Advantages and challenges in seed technology. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Gudkov, S.V.; Grinberg, M.A.; Sukhov, V.; Vodeneev, V. Effect of ionizing radiation on physiological and molecular processes in plants. J. Environ. Radioact. 2019, 202, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Volkova, P.Y.; Duarte, G.T.; Soubigou-Taconnat, L.; Kazakova, E.A.; Pateyron, S.; Bondarenko, V.S.; Bitarishvili, S.V.; Makarenko, E.S.; Churyukin, R.S.; Lychenkova, M.A.; et al. Early response of barley embryos to low- and high-dose gamma irradiation of seeds triggers changes in the transcriptional profile and an increase in hydrogen peroxide content in seedlings. J. Agron. Crop Sci 2020, 206, 277–295. [Google Scholar] [CrossRef]
- Volkova, P.Y.; Clement, G.; Makarenko, E.S.; Kazakova, E.A.; Bitarishvili, S.V.; Lychenkova, M.A. Metabolic profiling of γ-irradiated barley plants identifies reallocation of nitrogen metabolism and metabolic stress response. Dose-Response 2020, 18, 155932582091418. [Google Scholar] [CrossRef] [Green Version]
- Agathokleous, E.; Calabrese, E.J. Hormesis can enhance agricultural sustainability in a changing world. Glob. Food Secur. 2019, 20, 150–155. [Google Scholar] [CrossRef]
- De Micco, V.; Arena, C.; Pignalosa, D.; Durante, M. Effects of sparsely and densely ionizing radiation on plants. Radiat. Environ. Biophys. 2011, 50, 1–19. [Google Scholar] [CrossRef]
- Penfield, S. Seed dormancy and germination. Curr. Biol. 2017, 27, 874–878. [Google Scholar] [CrossRef] [Green Version]
- Geras’kin, S.; Churyukin, R.; Volkova, P. Radiation exposure of barley seeds can modify the early stages of plants’ development. J. Environ. Radioact. 2017, 177, 71–83. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleyer, M.; Trinogga, J.; Cebrián-Piqueras, M.A.; Trenkamp, A.; Fløjgaard, C.; Ejrnæs, R.; Bouma, T.J.; Minden, V.; Maier, M.; Mantilla-Contreras, J.; et al. Trait correlation network analysis identifies biomass allocation traits and stem specific length as hub traits in herbaceous perennial plants. J. Ecol. 2018. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Davidson, A. Adaptive phenotypic plasticity and plant water use. Funct. Plant Biol. 2010, 37, 117–127. [Google Scholar] [CrossRef]
- Giordano, M. Homeostasis: An underestimated focal point of ecology and evolution. Plant Sci. 2013, 211, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Mahapatra, K.; Roy, S. An insight into the mechanism of DNA damage response in plants- role of SUPPRESSOR OF GAMMA RESPONSE 1: An overview. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2020, 819–820, 111689. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Aoshima, N.; Takahashi, N.; Sakamoto, T.; Hiruma, K.; Saijo, Y.; Hidema, J.; Umeda, M.; Kimura, S. SUPPRESSOR OF GAMMA RESPONSE 1 acts as a regulator coordinating crosstalk between DNA damage response and immune response in Arabidopsis thaliana. Plant Mol. Biol. 2020, 103, 321–340. [Google Scholar] [CrossRef]
- Yao, S.; Luo, S.; Pan, C.; Xiong, W.; Xiao, D.; Wang, A.; Zhan, J.; He, L. Metacaspase MC1 enhances aluminum-induced programmed cell death of root tip cells in Peanut. Plant Soil 2020, 448, 479–494. [Google Scholar] [CrossRef]
- Coll, N.S.; Smidler, A.; Puigvert, M.; Popa, C.; Valls, M.; Dangl, J.L. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: Functional linkage with autophagy. Cell Death Differ. 2014, 21, 1399–1408. [Google Scholar] [CrossRef] [Green Version]
- Shamloo-Dashtpagerdi, R.; Lindlöf, A.; Aliakbari, M.; Pirasteh-Anosheh, H. Plausible association between drought stress tolerance of barley (Hordeum vulgare L.) and programmed cell death via MC1 and TSN1 genes. Physiol. Plant. 2020, 170, 46–59. [Google Scholar] [CrossRef]
- Caarls, L.; Elberse, J.; Awwanah, M.; Ludwig, N.R.; de Vries, M.; Zeilmaker, T.; Van Wees, S.C.M.; Schuurink, R.C.; Van den Ackerveken, G. Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc. Natl. Acad. Sci. USA 2017, 114, 6388–6393. [Google Scholar] [CrossRef] [Green Version]
- Rustgi, S.; Springer, A.; Kang, C.; von Wettstein, D.; Reinbothe, C.; Reinbothe, S.; Pollmann, S. ALLENE OXIDE SYNTHASE and HYDROPEROXIDE LYASE, two non-canonical cytochrome P450s in Arabidopsis thaliana and their different roles in plant defense. IJMS 2019, 20, 3064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Xu, W.; Deng, C.; Xu, S.; Li, F.; Wu, Y.; Wu, L.; Bian, P. A pivotal role of the jasmonic acid signal pathway in mediating radiation-induced bystander effects in Arabidopsis thaliana. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2016, 791–792, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Mithöfer, A.; Kombrink, E.; Boland, W.; Hamamoto, S.; Uozumi, N.; Tohma, K.; Ueda, M. 12-Hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in Samanea saman. Plant Physiol. 2011, 155, 1226–1236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krieger-Liszkay, A.; Shimakawa, G.; Sétif, P. Role of the two PsaE isoforms on O2 reduction at photosystem I in Arabidopsis thaliana. Biochim. Biophys. Acta (Bba) Bioenerg. 2020, 1861, 148089. [Google Scholar] [CrossRef]
- Sewelam, N.; Kazan, K.; Schenk, P.M. Global plant stress signaling: Reactive oxygen species at the cross-road. Front. Plant Sci. 2016, 7, 187. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress: ROS and redox signalling in plants. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, F.; Wu, Y.; Lou, L.; Liu, L.; Tian, M.; Ning, Y.; Shu, K.; Tang, S.; Xie, Q. The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 2015, 27, 214–227. [Google Scholar] [CrossRef] [Green Version]
- Voiniciuc, C.; Schmidt, M.H.-W.; Berger, A.; Yang, B.; Ebert, B.; Scheller, H.V.; North, H.M.; Usadel, B.; Günl, M. MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol. 2015, 169, 403–420. [Google Scholar] [CrossRef]
- Xu, D.; Huang, X.; Xu, Z.-Q.; Schläppi, M. The HyPRP gene EARLI1 has an auxiliary role for germinability and early seedling development under low temperature and salt stress conditions in Arabidopsis thaliana. Planta 2011, 234, 565–577. [Google Scholar] [CrossRef]
- Barrero, J.M.; Dorr, M.M.; Talbot, M.J.; Ishikawa, S.; Umezawa, T.; White, R.G.; Gubler, F. A role for PM19-Like 1 in seed dormancy in Arabidopsis. Seed Sci. Res. 2019, 29, 184–196. [Google Scholar] [CrossRef]
- Rerksiri, W.; Zhang, X.; Xiong, H.; Chen, X. Expression and promoter analysis of six heat stress-inducible genes in rice. Sci. World J. 2013, 2013, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lan, H.; Huang, P.; Zhang, Y.; Yuan, X.; Huang, X.; Huang, J.; Zhang, H. Characterization of OsPM19L1 encoding an AWPM-19-like family protein that is dramatically induced by osmotic stress in rice. Genet. Mol. Res. 2015, 14, 11994–12005. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.D.; Castillejo-Pons, P.; Alsaif, O.; Stahl, Y.; Seale, M.; Morris, P.C. The conserved plant PM19 protein functions as an osmosensor and regulator of germination. bioRxiv 2020. [Google Scholar] [CrossRef]
- Bitarishvili, S.V.; Volkova, P.Y.; Geras’kin, S.A. γ-Irradiation of barley seeds and its effect on the phytohormonal status of seedlings. Russ. J. Plant Physiol. 2018, 65, 446–454. [Google Scholar] [CrossRef]
- Yao, L.; Cheng, X.; Gu, Z.; Huang, W.; Li, S.; Wang, L.; Wang, Y.-F.; Xu, P.; Ma, H.; Ge, X. The AWPM-19 family protein OsPM1 mediates abscisic acid influx and drought response in rice. Plant Cell 2018, 30, 1258–1276. [Google Scholar] [CrossRef]
- Mangelsen, E.; Kilian, J.; Harter, K.; Jansson, C.; Wanke, D.; Sundberg, E. Transcriptome analysis of high-temperature stress in developing barley caryopses: Early stress responses and effects on storage compound biosynthesis. Mol. Plant 2011, 4, 97–115. [Google Scholar] [CrossRef]
- Barrero, J.M.; Cavanagh, C.; Verbyla, K.L.; Tibbits, J.F.G.; Verbyla, A.P.; Huang, B.E.; Rosewarne, G.M.; Stephen, S.; Wang, P.; Whan, A.; et al. Transcriptomic analysis of wheat near-isogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 2015, 16, 93. [Google Scholar] [CrossRef] [Green Version]
- Thor, K. Calcium—Nutrient and Messenger. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Munir, S.; Khan, M.; Song, J.; Munir, S.; Zhang, Y.; Ye, Z.; Wang, T. Genome-wide identification, characterization and expression analysis of calmodulin-like (CML) proteins in tomato (Solanum lycopersicum). Plant Physiol. Biochem. 2016, 102, 167–179. [Google Scholar] [CrossRef]
- Bender, K.W.; Rosenbaum, D.M.; Vanderbeld, B.; Ubaid, M.; Snedden, W.A. The Arabidopsis calmodulin-like protein, CML39, functions during early seedling establishment. Plant J. 2013, 76, 634–647. [Google Scholar] [CrossRef]
- Midhat, U.; Ting, M.K.Y.; Teresinski, H.J.; Snedden, W.A. The calmodulin-like protein, CML39, is involved in regulating seed development, germination, and fruit development in Arabidopsis. Plant Mol. Biol. 2018, 96, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Zhou, Q.; Chen, C.; Cui, Q.; Zhao, Y.; Wang, K.; Arkorful, E.; Chen, X.; Sun, K.; Li, X. Isolation and expression analysis of CsCML genes in response to abiotic stresses in the tea plant (Camellia sinensis). Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]


| № | Ensembl ID | Name of Homologues | Full Name | Biological Process |
|---|---|---|---|---|
| 1 | HORVU6Hr1G053540 | SOG1 | Suppressor of gamma response 1 | DNA repair |
| 2 | HORVU4Hr1G090300 | ATP1 | Probable pterin-4-alpha-carbinolamine dehydratase, chloroplastic | ABA signaling |
| 3 | HORVU0Hr1G016920 | HSP7R | Heat shock 70 kDa protein 17 | Chaperon activity |
| 4 | HORVU2Hr1G018440 | PER2 | Peroxidase 2 | Antioxidant process |
| 5 | HORVU6Hr1G071920 | GT7 | Probable glycosyltransferase 7 | Biosynthesis of cell wall |
| 6 | HORVU1Hr1G066540 | PER1 | Peroxidase 1 | Antioxidant process |
| 7 | HORVU7Hr1G046320 | CB121 | Chlorophyll a-b binding protein 1B-21, chloroplastic | Photosynthesis |
| 8 | HORVU4Hr1G013840 | DIOX6 | Probable 2-oxoglutarate-dependent dioxygenase | Anthocyanin biosynthesis |
| 9 | HORVU2Hr1G098860 | CIP1 | COP1-interactive protein 1 | ABA signaling |
| 10 | HORVU3Hr1G109230 | CML31 | Probable calcium-binding protein CML31 | Calcium signaling |
| 11 | HORVU3Hr1G095700 | MCA1 | Metacaspase-1 | Protein catabolism |
| 12 | HORVU7Hr1G056820 | HFB2B | Heat stress transcription factor B-2b | Chaperon activity |
| 13 | HORVU5Hr1G125450 | PM19L | Membrane protein PM19L | ABA signaling |
| 14 | HORVU7Hr1G117000 | ENPL | Endoplasmin homolog | Molecular chaperone |
| 15 | HORVU5Hr1G113900 | PSAE | Photosystem I reaction center subunit IV, chloroplastic | Photosynthesis |
| 16 | HORVU2Hr1G114680 | EARLI1 | Lipid transfer protein EARLI 1 | Lipid transport |
| 17 | HORVU4Hr1G066230 | AOS2 | Allene oxide synthase 2 | Jasmonic acid biosynthesis |
| Ensembl Name | Short Gene Name | Cultivar | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stimulation | No Effect | Inhibition | |||||||||||||
| Fox 1 | Ratnik | Eryoma | Master | Grees | Fedos | Leon | |||||||||
| R | S | R | S | R | S | R | S | R | S | R | S | R | S | ||
| HORVU6Hr1G053540 | SOG1 | −1.9 | Rad | ND | ND | Ref | ND | ND | Ref | ND | ND | −2.2 | ND | 2.1 | ND |
| HORVU4Hr1G090300 | ATP1 | 1.1 | ND | Ref | ND | 3.4 | −1.1 | −1.6 | 1.3 | Ref | Ref | −1.9 | Rad | 1.4 | 1.4 |
| HORVU0Hr1G016920 | HSP7R | 2.3 | 1.5 | 1.6 | 1.2 | 1.4 | −1.7 | −1.6 | −1.7 | −1.1 | 1.1 | 1.5 | −1.1 | 1.0 | −1.2 |
| HORVU2Hr1G018440 | PER2 | ND | ND | Rad | ND | ND | 2.2 | ND | ND | ND | ND | ND | 1.7 | Ref | Rad |
| HORVU6Hr1G071920 | GT7 | Ref | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | Rad | Ref | Rad |
| HORVU1Hr1G066540 | PER1 | ND | Ref | 1.3 | ND | ND | ND | ND | ND | −1.4 | ND | 1.7 | ND | 1.0 | ND |
| HORVU7Hr1G046320 | CB121 | ND | −1.1 | ND | 1.2 | ND | −2.2 | ND | 1.2 | ND | −1.1 | ND | −1.2 | ND | −1.4 |
| HORVU4Hr1G013840 | DIOX6 | 1.3 | 1.9 | 1.1 | −1.1 | 1.2 | 1.7 | 1.0 | −1.6 | -1.2 | 1.2 | 2.2 | −1.6 | 1.0 | Rad |
| HORVU2Hr1G098860 | CIP1 | 1.6 | 1.1 | 1.0 | 1.1 | 1.2 | −1.3 | 1.0 | −1.2 | 1.2 | 1.0 | 1.2 | −1.2 | −1.1 | 1.1 |
| HORVU3Hr1G109230 | CML31 | ND | ND | 8.7 | ND | 6.5 | Rad | Rad | ND | ND | ND | −1.1 | −1.4 | −2.9 | −1.1 |
| HORVU3Hr1G095700 | MCA1 | 1.9 | 1.1 | 1.1 | 1.2 | 1.5 | −1.8 | −1.7 | −1.1 | 1.7 | 1.1 | 2.7 | −1.5 | 1.9 | 1.5 |
| HORVU7Hr1G056820 | HFB2B | Ref | Rad | ND | ND | ND | ND | Ref | Rad | ND | ND | Ref | ND | ND | ND |
| HORVU5Hr1G125450 | PM19L | 1.1 | 14.9 | 3.8 | −1.6 | 1.9 | 5.2 | −1.9 | −9.5 | 2.8 | 2.5 | 2.1 | −1.2 | −1.6 | 2.2 |
| HORVU7Hr1G117000 | ENPL | 1.8 | 1.4 | 1.3 | 1.6 | 1.5 | −1.8 | −1.4 | −1.6 | −1.1 | −1.1 | 1.5 | −1.4 | 1.0 | 1.0 |
| HORVU5Hr1G113900 | PSAE | 1.2 | 1.1 | ND | 1.2 | 3.0 | −2.3 | 1.4 | 1.3 | ND | 1.0 | Rad | −1.2 | ND | −1.3 |
| HORVU2Hr1G114680 | EARLI1 | ND | Ref | ND | ND | ND | Rad | Ref | 1.2 | ND | Rad | ND | 1.2 | ND | Rad |
| HORVU4Hr1G066230 | AOS2 | 3.2 | 1.1 | 3.3 | 1.1 | 1.5 | −2.2 | 1.7 | −1.3 | 1.2 | 1.0 | Ref | −1.1 | −1.5 | 1.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorbatova, I.V.; Kazakova, E.A.; Podlutskii, M.S.; Pishenin, I.A.; Bondarenko, V.S.; Dontsova, A.A.; Dontsov, D.P.; Snegirev, A.S.; Makarenko, E.S.; Bitarishvili, S.V.; et al. Studying Gene Expression in Irradiated Barley Cultivars: PM19L-like and CML31-like Expression as Possible Determinants of Radiation Hormesis Effect. Agronomy 2020, 10, 1837. https://doi.org/10.3390/agronomy10111837
Gorbatova IV, Kazakova EA, Podlutskii MS, Pishenin IA, Bondarenko VS, Dontsova AA, Dontsov DP, Snegirev AS, Makarenko ES, Bitarishvili SV, et al. Studying Gene Expression in Irradiated Barley Cultivars: PM19L-like and CML31-like Expression as Possible Determinants of Radiation Hormesis Effect. Agronomy. 2020; 10(11):1837. https://doi.org/10.3390/agronomy10111837
Chicago/Turabian StyleGorbatova, Irina V., Elizaveta A. Kazakova, Mikhail S. Podlutskii, Ivan A. Pishenin, Vladimir S. Bondarenko, Aleksandra A. Dontsova, Dmitriy P. Dontsov, Aleksei S. Snegirev, Ekaterina S. Makarenko, Sofia V. Bitarishvili, and et al. 2020. "Studying Gene Expression in Irradiated Barley Cultivars: PM19L-like and CML31-like Expression as Possible Determinants of Radiation Hormesis Effect" Agronomy 10, no. 11: 1837. https://doi.org/10.3390/agronomy10111837
APA StyleGorbatova, I. V., Kazakova, E. A., Podlutskii, M. S., Pishenin, I. A., Bondarenko, V. S., Dontsova, A. A., Dontsov, D. P., Snegirev, A. S., Makarenko, E. S., Bitarishvili, S. V., Lychenkova, M. A., Chizh, T. V., & Volkova, P. Y. (2020). Studying Gene Expression in Irradiated Barley Cultivars: PM19L-like and CML31-like Expression as Possible Determinants of Radiation Hormesis Effect. Agronomy, 10(11), 1837. https://doi.org/10.3390/agronomy10111837