Expansion of Native Plant Stellera chamaejasme L. Alters the Structure of Soil Diazotrophic Community in a Salinized Meadow Grassland, Northeast China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Location and Site Description
2.2. Experimental Design and Soil Sampling
2.3. Soil Physicochemical Property Measurements
2.4. Soil DNA Extraction
2.5. nifH Gene Sequencing and Bioinformatics Analysis
2.6. Data Analysis
3. Results
3.1. Soil Physicochemical Properties
3.2. Alpha Diversity and Taxonomic Composition
3.3. Correlation Analysis between the Relative Abundances of Taxonomic Groups and Soil Physicochemical Properties
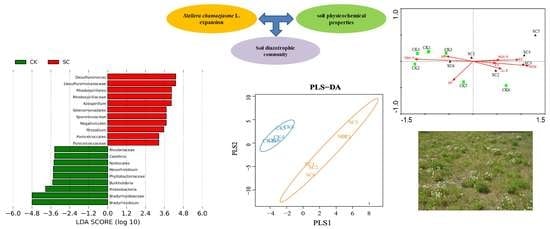
3.4. Biomarkers for SC and CK
3.5. Diazotrophic Structure and Correlation with Soil Physicochemical Properties
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valéry, L.; Fritz, H.; Lefeuvre, J.C.; Simberloff, D. In search of a real definition of the biological invasion phenomenon itself. Biol. Invasions 2008, 10, 1345–1351. [Google Scholar] [CrossRef]
- Wilson, J.R.U.; Dormontt, E.E.; Prentis, P.J.; Lowe, A.J.; Richardson, D.M. Something in the way you move: Dispersal pathways affect invasion success. Trends Ecol. Evol. 2009, 24, 136–144. [Google Scholar] [CrossRef]
- Canals, R.; Pedro, J.; Rupérez, E.; San-Emeterio, L. Nutrient pulses after prescribed winter fires and preferential patterns of N uptake may contribute to the expansion of Brachypodium pinnatum (L.) P. Beauv. in highland grasslands. Appl. Veg. Sci. 2014, 17, 419–428. [Google Scholar] [CrossRef]
- Muñoz-Vallés, S.; Cambrollé, J. The threat of native-invasive plant species to biodiversity conservation in coastal dunes. Ecol. Eng. 2015, 79, 32–34. [Google Scholar] [CrossRef]
- Torell, L.A.; Mcdaniel, K.C.; Brown, J.R.; Torell, G.L. Broom snakeweed (Gutierrezia sarothrae) population change in central New Mexico: Implications for management and control. Rangel. Ecol. Manag. 2018, 71, 228–238. [Google Scholar] [CrossRef]
- Vilà, M.; Espinar, J.L.; Hejda, M.; Hulme, P.E.; Jarosik, V.; Maron, J.L.; Pergl, J.; Schaffner, U.; Sun, Y.; Pysek, P. Ecological impacts of invasive alien plants: A meta-analysis of their effects on species, communities and ecosystems. Ecol. Lett. 2011, 14, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Carey, M.P.; Sanderson, B.L.; Barnas, K.A.; Olden, J.D. Native invaders-challenges for science management, policy, and society. Front. Ecol. Environ. 2012, 10, 373–381. [Google Scholar] [CrossRef]
- Dassonville, N.; Vanderhoeven, S.; Vanparys, V.; Hayez, M.; Gruber, W.; Meerts, P. Impacts of alien invasive plants on soil nutrients are correlated with initial site conditions in NW Europe. Oecologia 2008, 157, 131–140. [Google Scholar] [CrossRef]
- Fan, L.; Chen, Y.; Yuan, J.G.; Yang, Z.Y. The effect of Lantana camara Linn. invasion on soil chemical and microbiological properties and plant biomass accumulation in southern China. Geoderma 2010, 154, 370–378. [Google Scholar] [CrossRef]
- Kumar, M.; Verma, A.K.; Garkoti, S.C. Lantana camara and Ageratina adenophora invasion alter the understory species composition and diversity of chir pine forest in central Himalaya, India. Acta Oecol. 2010, 109, 103642. [Google Scholar] [CrossRef]
- Ruwanza, S. Effects of Solanum mauritianum Scopoli (bugweed) invasion on soil and vegetation in Vhembe Biosphere Reserve, South Africa. Austral Ecology 2021, 46, 342–348. [Google Scholar] [CrossRef]
- Chen, D.; Xing, W.; Lan, Z.; Saleem, M.; Saleem, M.; Wu, Y.; Hu, S.; Bai, Y. Direct and indirect effects of nitrogen enrichment on soil organisms and carbon and nitrogen mineralization in a semi-arid grassland. Funct. Ecol. 2019, 33, 175–187. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Yan, Y.E.; Jiang, F.; Leng, X.; Cheng, X.L.; An, S.Q. Response of the soil microbial community composition and biomass to a short-term Spartina alterniflora invasion in a coastal wetland of eastern China. Plant Soil 2016, 408, 443–456. [Google Scholar] [CrossRef]
- Stefanowicz, A.M.; Zubek, S.; Stanek, M.; Grzes, L.M.; Rozej-Pabijan, E.; Blaszkowski, J.; Woch, M.W. Invasion of Rosa rugosa induced changes in soil nutrients and microbial communities of coastal sand dunes. Sci. Total Environ. 2019, 677, 340–349. [Google Scholar] [CrossRef]
- Batten, K.M.; Scow, K.M.; Davies, K.F.; Harrison, S.P. Two invasive plants alter soil microbial community composition in serpentine grasslands. Biol. Invasions 2006, 8, 217–230. [Google Scholar] [CrossRef]
- Souza-Alonso, P.; Novoa, A.; González, L. Soil biochemical alterations and microbial community responses under Acacia dealbata Link invasion. Soil Biol. Biochem. 2014, 79, 100–108. [Google Scholar] [CrossRef]
- Rodríguez-Caballero, G.; Caravaca, F.; Alguacil, M.M.; Fernández-López, M.; Fernández-González, A.J.; Roldán, A. Striking alterations in the soil bacterial community structure and functioning of the biological N cycle induced by Pennisetum setaceum invasion in a semiarid environment. Soil Biol. Biochem. 2017, 109, 176–187. [Google Scholar] [CrossRef]
- Zhao, M.X.; Lu, X.F.; Zhao, H.X.; Yang, Y.F.; Hale, L.; Gao, Q.; Liu, W.X.; Guo, J.Y.; Li, Q.; Zhou, J.Z.; et al. Ageratina adenophora invasions are associated with microbially mediated differences in biogeochemical cycles. Sci. Total Environ. 2019, 677, 47–56. [Google Scholar] [CrossRef]
- Xu, M.P.; Wang, J.Y.; Han, X.H.; Ren, C.J.; Yang, G.H. Plant biomass and soil nutrients mainly explain the variation of soil microbial communities during secondary succession on the Loess Plateau. Microb. Ecol. 2021. [Google Scholar] [CrossRef]
- Lau, J.A.; Suwa, T. The changing nature of plant–microbe interactions during a biological invasion. Biol. Invasions 2016, 18, 3527–3534. [Google Scholar] [CrossRef]
- Novoa, A.; Rodríguez, R.; Richardson, D.; Gonzalez, L. Soil quality: A key factor in understanding plant invasion? The case of Carpobrotus edulis (L.) N.E.Br. Biol. Invasions 2014, 16, 429–443. [Google Scholar] [CrossRef]
- Yahdjian, L.; Gherardim, L.; Sala, O.E. Nitrogen limitation in arid-subhumid ecosystems: A meta-analysis of fertilization studies. J. Arid Environ. 2011, 75, 675–680. [Google Scholar] [CrossRef]
- Zong, N.; Song, M.H.; Zhao, G.S.; Shi, P.L. Nitrogen economy of alpine plants on the north Tibetan Plateau: Nitrogen conservation by resorption rather than open sources through biological symbiotic fixation. Ecol. Evol. 2020, 10, 2051–2061. [Google Scholar] [CrossRef]
- Chen, F.S.; Zeng, D.H.; Fahey, T.J. Changes in soil nitrogen availability due to stand development and management practices on semi-arid sandy lands, in northern China. Land Degrad. Dev. 2010, 20, 481–491. [Google Scholar] [CrossRef]
- Yu, Q.; Wilcox, K.; La Pierre, K.; Knapp, A.K.; Han, X.G.; Smith, M.D. Stoichiometric homeostasis predicts plant species dominance, temporal stability, and responses to global change. Ecology 2015, 96, 2328–2335. [Google Scholar] [CrossRef] [PubMed]
- Hurst, A.; John, E. The biotic and abiotic changes associated with Brachypodium pinnatum dominance in chalk grassland in south–east England. Biol. Conserv. 1999, 88, 75–84. [Google Scholar] [CrossRef]
- Fraterrigo, J.M.; Strickland, M.S.; Keiser, A.D. Bradford, M.A. Nitrogen uptake and preference in a forest understory following invasion by an exotic grass. Oecologia 2011, 167, 781–791. [Google Scholar] [CrossRef]
- Ribeiro, P.C.D.; Menendez, E.; da Silva, D.L.; Bonieck, D.; Ramirez-Bahena, M.H.; Resende-Stoianoff, M.A. Peix, A.; Velazquez, E.; Mateos, P.F.; Scotti, M.R. Invasion of the Brazilian campo rupestre by the exotic grass Melinis minutiflora is driven by the high soil N availability and changes in the N cycle. Sci. Total Environ. 2017, 577, 202–211. [Google Scholar] [CrossRef]
- Chmolowska, D.; Nobis, M.; Nowak, A.; Maslak, M.; Kojs, P.; Rutkowska, J.; Zubek, S. Rapid change in forms of inorganic nitrogen in soil and moderate weed invasion following translocation of wet meadows to reclaimed post-industrial land. Land Degrad. Dev. 2019, 30, 964–978. [Google Scholar] [CrossRef]
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.C.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Shen, W.J.; Xu, H.; Li, Y.D.; Luo, T.S. The composition of nitrogen-fixing microorganisms correlates with soil nitrogen content during reforestation: A comparison between legume and non-legume plantations. Front. Microbiol. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.Y.; Zhou, J.W.; Liu, J.; Jiang, K.; Du, D.L. Responses of soil N-fixing bacteria communities to Amaranthus retroflexus invasion under different forms of N deposition. Agric. Ecosyst. Environ. 2017, 247, 329–336. [Google Scholar] [CrossRef]
- Feng, B.M.; Pei, Y.H.; Hua, H.M. Chemical constituents of Stellera chamaejasme L. J. Asian Nat. Prod. Res. 2002, 4, 259–263. [Google Scholar] [CrossRef]
- Yan, Z.Q.; Guo, H.R.; Yang, J.Y.; Liu, Q.; Jin, H.; Xu, R.; Cui, H.Y.; Qin, B. Phytotoxic flavonoids from roots of Stellera chamaejasme L. (Thymelaeaceae). Phytochemistry. 2014, 106, 61–68. [Google Scholar] [CrossRef]
- Xing, F.; Guo, J.X.; Wang, Y.H. Seed germination characteristics and regeneration mechanism of Stellera chamaejasme population. Chin. J. Appl. Ecol. 2003, 14, 1851–1854, (In English Abstract). [Google Scholar]
- Xing, F.; Wang, Y.H.; Guo, J.X. Spatial distribution patterns and dispersal mechanisms of the seed population of Stellera chamaejasme on degraded grasslands in Inner Mongolia. Chin. Acta Ecol. Sin. 2004, 24, 143–148, (In English Abstract). [Google Scholar]
- Sun, G.; Luo, P.; Wu, N.; Qiu, P.F.; Gao, Y.H.; Chen, H.; Shi, F.S. Stellera chamaejasme L. increases soil N availability, turnover rates and microbial biomass in an alpine meadow ecosystem on the eastern Tibetan Plateau of China. Soil Biol. Biochem. 2009, 41, 86–91. [Google Scholar] [CrossRef]
- Li, S.G.; Harazono, Y.; Oikawa, T.; Zhao, H.L.; Chang, X.L. Grassland desertification by grazing and the resulting micrometeorological changes in Inner Mongolia. Agric. For. Meteorol. 2000, 102, 125–137. [Google Scholar] [CrossRef]
- Zhang, Y.; Cao, C.Y.; Guo, L.N.; Wu, Q.; Cui, Z.B. Soil properties, bacterial community composition, and metabolic diversity responses to soil salinization of a semiarid grassland in northeast Chinese. J. Soil Water Conserv. 2015, 70, 110–120. [Google Scholar] [CrossRef]
- Cao, C.Y.; Jiang, D.M.; Zhu, L.H.; Nan, Y.H. Degradation and diversity changes of meadow grassland in Keerqin Sandy Land. Acta Prataculturae Sin. 2006, 15, 8–26, (In English Abstract). [Google Scholar]
- Institute of Soil Science, Chinese Academy of Sciences (ISSCAS). Methods on Soil Microorganisms Study; Science Press: Beijing, China, 1978. [Google Scholar]
- Poly, F.; Ranjard, L.; Nazaret, S.; Gourbiere, F.; Monrozier, L.J. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 2001, 67, 2255–2262. [Google Scholar] [CrossRef] [Green Version]
- Gaby, J.C.; Buckley, D.H. A comprehensive aligned nifH gene database: A multipurpose tool for studies of nitrogen-fixing bacteria. Database 2014, bau001. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Wu, H.; Ye, J.Y.; Zhong, B. Study on the release routes of allelochemicals from Pistia stratiotes Linn., and its anti-cyanobacteria mechanisms on Microcystis aeruginosa. Environ. Sci. Pollut. Res. 2015, 22, 18994–19001. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, W.L.; Zhang, C.B.; Wang, J. Effects of Solidago canadensis invadation on dynamics of native plant communities and their mechanisms. Chin. J. Plant. Eco. 2012, 36, 253–261, (In English Abstract). [Google Scholar] [CrossRef]
- Huangfu, C.H.; Li, H.Y.; Chen, X.W.; Liu, H.M.; Wang, H.; Yang, D.L. Response of an invasive plant, Flaveria bidentis, to nitrogen addition: A test of form-preference uptake. Biol. Invasions 2016, 18, 3365–3380. [Google Scholar] [CrossRef]
- Valéry, L.; Bouchard, V.; Lefeuvre, J.C. Impact of the invasive native species Elymus athericus on carbon pools in a salt marsh. Wetlands 2004, 24, 268–276. [Google Scholar] [CrossRef]
- James, J.J. Leaf nitrogen productivity as a mechanism driving the success of invasive annual grasses under low and high nitrogen supply. J. Arid. Environ. 2008, 72, 1775–1784. [Google Scholar] [CrossRef]
- Wang, Y.S.; Li, C.N.; Kou, Y.P.; Wang, J.J.; Tu, B.; Li, H.; Li, X.Z.; Wang, C.T.; Yao, M.J. Soil pH is a major driver of soil diazotrophic community assembly in Qinghai-Tibet alpine meadows. Soil Biol. Biochem. 2017, 115, 547–555. [Google Scholar] [CrossRef]
- Liang, Y.M.; Pan, F.J.; He, X.Y.; Chen, X.B.; Su, Y.R. Effect of vegetation types on soil arbuscular mycorrhizal fungi and nitrogen-fixing bacterial communities in a karst region. Environ. Sci. Pollut. Res. 2016, 23, 18482–18491. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.K.; Weisenhorn, P.; Gilbert, J.A.; Shi, Y.; Bai, Y.; Chu, H.Y. Soil pH correlates with the co-occurrence and assemblage process of diazotrophic communities in rhizosphere and bulk soils of wheat fields. Soil Biol. Biochem. 2018, 121, 185–192. [Google Scholar] [CrossRef]
- Han, Z.M.; Deng, M.W.; Yuan, A.Q.; Wang, J.H.; Li, H.; Ma, J.C. Vertical variation of a black soil’s properties in response to freeze-thaw cycles and its links to shift of microbial community structure. Sci. Total Environ. 2018, 625, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Y.; Wang, Q.; Ouyang, H.L.; Lan, S.B.; Hu, C.X. Pyrosequencing reveals significant changes in microbial communities along the ecological succession of biological soil crusts in the Tengger Desert of China. Pedosphere 2018, 28, 350–362. [Google Scholar] [CrossRef]
- Bui, E.N. Soil salinity: A neglected factor in plant ecology and biogeography. J. Arid. Environ. 2013, 92, 4–25. [Google Scholar] [CrossRef]
- Xu, Y.D.; Wang, T.; Li, H.; Ren, C.J.; Chen, J.W.; Yang, G.H.; Han, X.H.; Feng, Y.Z.; Ren, G.X.; Wang, X.J. Variations of soil nitrogen-fixing microorganism communities and nitrogen fractions in a Robinia pseudoacacia chronosequence on the Loess Plateau of China. Catena 2019, 174, 316–323. [Google Scholar] [CrossRef]
- Martínez, J.M.; Galantini, J.A.; Duval, M.E.; López, F.M. Tillage effects on labile pools of soil organic nitrogen in a semi-humid climate of Argentina: A long-term field study. Soil Tillage Res. 2017, 169, 71–80. [Google Scholar] [CrossRef]
- Qiang, W.; He, L.; Zhang, Y.; Liu, B.; Pang, X. Aboveground vegetation and soil physicochemical properties jointly drive the shift of soil microbial community during subalpine secondary succession in southwest China. Catena 2021, 202, 105251. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G. Effects of exotic plant invasions on soil nutrient cycling processes. Ecosystems 2003, 6, 503–523. [Google Scholar] [CrossRef]
- Inderjit; van der Putten, W.H. Impacts of soil microbial communities on exotic plant invasions. Trends Ecol. Evol. 2010, 25, 512–519. [Google Scholar] [CrossRef]
- Lindsay, E.A.; Colloff, M.J.; Gibb, N.L.; Wakelin, S.A. The abundance of microbial functional genes in grassy woodlands is influenced more by soil nutrient enrichment than by recent weed invasion or livestock exclusion. Appl. Environ. Microbiol. 2010, 76, 5547–5555. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.W.; Yang, M.Z.; Chen, Y.J.; Chen, L.M.; Zhang, D.Z.; Mei, L.; Shi, Y.T.; Zhang, H.B. Changes in non-symbiotic nitrogen-fixing bacteria inhabiting rhizosphere soils of an invasive plant Ageratina adenophora. Appl. Soil Ecol. 2012, 54, 32–38. [Google Scholar] [CrossRef]
- Wang, C.Y.; Jiang, K.; Zhou, J.W.; Wu, B.D. Solidago canadensis invasion affects soil N-fixing bacterial communities in heterogeneous landscapes in urban ecosystems in East China. Sci. Total Environ. 2018, 631–632, 702–713. [Google Scholar] [CrossRef]
- Le Roux, J.J.; Ellis, A.G.; van Zyl, L.-M.; Hosking, N.D.; Keet, J.H.; Yannelli, F.A. Importance of soil legacy effects and successful mutualistic interactions during Australian acacia invasions in nutrient poor environments. J. Ecol. 2018, 106, 2071–2081. [Google Scholar] [CrossRef]
- Puozaa, D.K.; Jaiswal, S.K.; Dakora, F.D. Phylogeny and distribution of Bradyrhizobium symbionts nodulating cowpea (Vigna unguiculata L. Walp) and their association with the physicochemical properties of acidic african soils. Syst. Appl. Microbiol. 2019, 42, 403–414. [Google Scholar] [CrossRef]
- Izquierdo, J.A.; Nusslein, K. Variation in diazotrophic community structure in forest soils reflects land use history. Soil Biol. Biochem. 2015, 80, 1–8. [Google Scholar] [CrossRef]
- Pierra, M.; Carmona-Martínez, A.A.; Trably, E.; Godon, J.J.; Bernet, N. Specific and efficient electrochemical selection of Geoalkalibacter subterraneus and Desulfuromonas acetoxidans in high current-producing biofilms. Bioelectrochemistry 2015, 106, 221–225. [Google Scholar] [CrossRef]





| Items | SC | CK | F | p |
|---|---|---|---|---|
| SOM (g·kg−1) | 3.83 ± 0.29a | 2.43 ± 0.88b | 13.659 | <0.01 |
| Total N (g·kg−1) | 0.91 ± 0.09a | 0.79 ± 0.07b | 6.357 | 0.03 |
| NH4+-N (mg·kg−1) | 1.74 ± 0.25a | 2.15 ± 0.25b | 8.199 | 0.017 |
| NO3−-N (mg·kg−1) | 0.82 ± 0.66a | 0.09 ± 0.08b | 7.319 | 0.022 |
| Total P (g·kg−1) | 0.52 ± 0.09a | 0.61 ± 0.07a | 3.58 | >0.05 |
| Available P (mg·kg−1) | 5.33 ± 0.93a | 5.25 ± 0.61a | 0.028 | >0.05 |
| pH | 8.125 ± 0.137a | 8.395 ± 0.227b | 6.233 | 0.032 |
| EC (µs·cm−1) | 251.8 ± 19.9a | 199.3 ± 48.6b | 5.983 | 0.034 |
| Samples | OTU Number Richness | ACE | Chao 1 | Shannon |
|---|---|---|---|---|
| CK | 2176.0 ± 172.9 | 2577.6 ± 381.2 | 2457.7 ± 329.3 | 9.6 ± 0.2 |
| SC | 2286.1 ± 376.3 | 2752.4 ± 739.5 | 2624.3 ± 637.1 | 9.7 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Cui, Z.; Wang, T.; Cao, C. Expansion of Native Plant Stellera chamaejasme L. Alters the Structure of Soil Diazotrophic Community in a Salinized Meadow Grassland, Northeast China. Agronomy 2021, 11, 2085. https://doi.org/10.3390/agronomy11102085
Zhang Y, Cui Z, Wang T, Cao C. Expansion of Native Plant Stellera chamaejasme L. Alters the Structure of Soil Diazotrophic Community in a Salinized Meadow Grassland, Northeast China. Agronomy. 2021; 11(10):2085. https://doi.org/10.3390/agronomy11102085
Chicago/Turabian StyleZhang, Ying, Zhenbo Cui, Tingting Wang, and Chengyou Cao. 2021. "Expansion of Native Plant Stellera chamaejasme L. Alters the Structure of Soil Diazotrophic Community in a Salinized Meadow Grassland, Northeast China" Agronomy 11, no. 10: 2085. https://doi.org/10.3390/agronomy11102085
APA StyleZhang, Y., Cui, Z., Wang, T., & Cao, C. (2021). Expansion of Native Plant Stellera chamaejasme L. Alters the Structure of Soil Diazotrophic Community in a Salinized Meadow Grassland, Northeast China. Agronomy, 11(10), 2085. https://doi.org/10.3390/agronomy11102085