microRNAs Mediated Regulation of the Ribosomal Proteins and its Consequences on the Global Translation of Proteins
Abstract
:1. Introduction
2. Materials and Methods
3. RPs on the Biogenesis and Assembly of Ribosomes, and Translation of Proteins
4. RPs Mediated Regulation of Biological Processes and Progression of Diseases
5. MicroRNAs Mediated Regulation of Gene Expression and Progression of Diseases
6. MicroRNAs Biogenesis and Dissemination to the Circulatory System
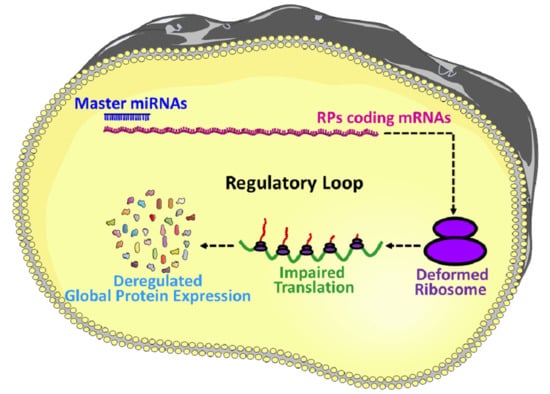
7. MicroRNAs in the Regulation of Ribosomal Protein Coding mRNAs
8. Conclusions
- I.
- The role of miRNAs in the regulation of gene expression is a well-investigated area of research, however, the roles of miRNAs in the regulation of RPs coding gene expression remains unexplored, and therefore, this area is required to be investigated further for a proper understanding of RPs synthesis, ribosomal assembly and regulation of global protein translation.
- II.
- The idea of master miRNAs that can influence the global translation of proteins is potentially true, and the existence of such miRNAs is further assured by the report of Orom et al. 2008, which showed that miR-10a binds to the 5′UTR of the RPs and regulates the global protein synthesis. However, further investigations are required to establish it as a scientific fact.
- III.
- RPs are an integral part of the translation machinery, required for the proper assembly and functioning of the ribosomes. Therefore, the ultimate results of the miRNA mediated regulation of the RPs are improper functioning of the translation machinery and deregulated synthesis proteins.
- IV.
- Ribosomopathy refers to a group of diseases caused by the deformed translation machinery, and many RPs are directly involved with ribosomopathy, thus finding the regulatory interaction of miRNAs and RPs could explore the regulatory role of miRNAs on ribosomopathy and might help to develop future therapeutic strategies.
- V.
- Deregulation of both RPs and miRNAs is very common in diseases including almost all types of cancers. Investigation of the miRNAs mediated regulation of RPs could provide a reasonable explanation behind the pathological conditions of these diseases.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ban, N.; Beckmann, R.; Cate, J.H.; Dinman, J.D.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 2014, 24, 165–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wool, I.G. The Structure and Function of Eukaryotic Ribosomes. Annu. Rev. Biochem. 1979, 48, 719–754. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Barna, M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell. Biol. 2012, 13, 355–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genuth, N.R.; Barna, M. Heterogeneity and specialized functions of translation machinery: From genes to organisms. Nat. Rev. Genet. 2018, 19, 431–452. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell. 2018, 71, 364–374. [Google Scholar] [CrossRef] [Green Version]
- Hausser, J.; Syed, A.P.; Bilen, B.; Zavolan, M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013, 23, 604–615. [Google Scholar] [CrossRef] [Green Version]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005, 120, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Reza, A.M.M.T.; Choi, Y.J.; Yuan, Y.G.; Das, J.; Yasuda, H.; Kim, J.H. MicroRNA-7641 is a regulator of ribosomal proteins and a promising targeting factor to improve the efficacy of cancer therapy. Sci. Rep. 2017, 7, 8365. [Google Scholar] [CrossRef] [Green Version]
- Ni, W.J.; Leng, X.M. miRNA-dependent activation of mRNA translation. MicroRNA 2016, 5, 83–86. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell. Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macfarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell. Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [Green Version]
- Dalmay, T. Mechanism of miRNA-mediated repression of mRNA translation. Essays Biochem. 2013, 54, 29–38. [Google Scholar]
- Cannell, I.G.; Kong, Y.W.; Bushell, M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008, 36, 1224–1231. [Google Scholar] [CrossRef] [Green Version]
- Chaillou, T.; Kirby, T.J.; McCarthy, J.J. Ribosome biogenesis: Emerging evidence for a central role in the regulation of skeletal muscle mass. J. Cell. Physiol. 2014, 229, 1584–1594. [Google Scholar] [CrossRef] [Green Version]
- Lempiainen, H.; Shore, D. Growth control and ribosome biogenesis. Curr. Opin. Cell. Biol. 2009, 21, 855–863. [Google Scholar] [CrossRef]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, C.G.; Teixeira, F.K.; Czech, B.; Preall, J.B.; Zamparini, A.L.; Seifert, J.R.; Malone, C.D.; Hannon, G.J.; Lehmann, R. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell. Stem. Cell. 2016, 18, 276–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebright, R.Y.; Lee, S.; Wittner, B.S.; Niederhoffer, K.L.; Nicholson, B.T.; Bardia, A.; Truesdell, S.; Wiley, D.F.; Wesley, B.; Li, S.; et al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science. 2020, 367, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Nag, S.; Zhang, X.; Wang, M.H.; Wang, H.; Zhou, J.; Zhang, R. Ribosomal proteins and human diseases: Pathogenesis, molecular mechanisms, and therapeutic implications. Med. Res. Rev. 2015, 35, 225–285. [Google Scholar] [CrossRef] [PubMed]
- Thomson, E.; Ferreira-Cerca, S.; Hurt, E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013, 126, 4815–4821. [Google Scholar] [CrossRef] [Green Version]
- Kressler, D.; Hurt, E.; Baβler, J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010, 1803, 673–683. [Google Scholar] [CrossRef] [Green Version]
- Nikolay, R.; van den Bruck, D.; Achenbach, J.; Nierhaus, K.H. Ribosomal proteins: Role in ribosomal functions. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2015. [Google Scholar]
- Martin-Marcos, P.; Hinnebusch, A.G.; Tamame, M. Ribosomal protein L33 is required for ribosome biogenesis, subunit joining, and repression of GCN4 translation. Mol. Cell. Biol. 2007, 27, 5968–5985. [Google Scholar] [CrossRef] [Green Version]
- Murguia, J.R.; Serrano, R. New functions of protein kinase Gcn2 in yeast and mammals. IUBMB Life 2012, 64, 971–974. [Google Scholar] [CrossRef]
- Tobin, C.; Mandava, C.S.; Ehrenberg, M.; Andersson, D.I.; Sanyal, S. Ribosomes lacking protein S20 are defective in mRNA binding and subunit association. J. Mol. Biol. 2010, 397, 767–776. [Google Scholar] [CrossRef]
- Moritz, M.; Paulovich, A.G.; Tsay, Y.F.; Woolford, J.L., Jr. Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J. Cell Biol. 1990, 111, 2261–2674. [Google Scholar] [CrossRef]
- Deshmukh, M.; Tsay, Y.F.; Paulovich, A.G.; Woolford, J.L., Jr. Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 1993, 13, 2835–2845. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, M.; Stark, J.; Yeh, L.C.; Lee, J.C.; Woolford, J.L., Jr. Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5S rRNA and assembly into ribosomes. J. Biol. Chem. 1995, 270, 30148–30156. [Google Scholar] [PubMed] [Green Version]
- Naganathan, A.; Wood, M.P.; Moore, S.D. The large ribosomal subunit protein L9 enables the growth of EF-P deficient cells and enhances small subunit maturation. PLoS ONE 2015, 10, e0120060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira-Cerca, S.; Pöll, G.; Gleizes, P.E.; Tschochner, H.; Milkereit, P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005, 20, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Takyar, S.; Hickerson, R.P.; Noller, H.F. mRNA helicase activity of the ribosome. Cell 2005, 120, 49–58. [Google Scholar] [CrossRef] [Green Version]
- Kramer, G.; Rauch, T.; Rist, W.; Vorderwülbecke, S.; Patzelt, H.; Schulze-Specking, A.; Ban, N.; Deuerling, E.; Bukau, B. L23 protein functions as a chaperone docking site on the ribosome. Nature 2002, 419, 171–174. [Google Scholar] [CrossRef]
- Pool, M.R.; Stumm, J.; Fulga, T.A.; Sinning, I.; Dobberstein, B. Distinct modes of signal recognition particle interaction with the ribosome. Science 2002, 297, 1345–1348. [Google Scholar] [CrossRef]
- Beckmann, R.; Spahn, C.M.; Eswar, N.; Helmers, J.; Penczek, P.A.; Sali, A.; Frank, J.; Blobel, G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 2001, 107, 361–372. [Google Scholar] [CrossRef] [Green Version]
- Clemons, W.M.; Ménétret, J.F.; Akey, C.W.; Rapoport, T.A. Structural insight into the protein translocation channel. Curr. Opin. Struct. Biol. 2004, 14, 390–396. [Google Scholar] [CrossRef]
- Alksne, L.E.; Anthony, R.A.; Liebman, S.W.; Warner, J.R. An accuracy center in the ribosome conserved over 2 billion years. Proc. Natl. Acad. Sci. USA 1993, 90, 9538–9541. [Google Scholar] [CrossRef] [Green Version]
- Stansfield, I.; Jones, K.M.; Herbert, P.; Lewendon, A.; Shaw, W.V.; Tuite, M.F. Missense translation errors in Saccharomyces cerevisiae. J. Mol. Biol. 1998, 282, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Synetos, D.; Frantziou, C.P.; Alksne, L.E. Mutations in yeast ribosomal proteins S28 and S4 affect the accuracy of translation and alter the sensitivity of the ribosomes to paromomycin. Biochim. Biophys. Acta 1996, 1309, 156–166. [Google Scholar] [CrossRef]
- Rother, S.; Strasser, K. The RNA polymerase II CTD kinase Ctk1 functions in translation elongation. Genes Dev. 2007, 21, 1409–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peltz, S.W.; Hammell, A.B.; Cui, Y.; Yasenchak, J.; Puljanowski, L.; Dinman, J.D. Ribosomal protein L3 mutants alter translational fidelity and promote rapid loss of the yeast killer virus. Mol. Cell. Biol. 1999, 19, 384–391. [Google Scholar] [CrossRef] [Green Version]
- Dresios, J.; Panopoulos, P.; Suzuki, K.; Synetos, D. A dispensable yeast ribosomal protein optimizes peptidyltransferase activity and affects translocation. J. Biol. Chem. 2003, 278, 3314–3322. [Google Scholar] [CrossRef] [Green Version]
- Meskauskas, A.; Dinman, J.D. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA 2001, 7, 1084–1096. [Google Scholar] [CrossRef] [Green Version]
- Gadal, O.; Strauss, D.; Kessl, J.; Trumpower, B.; Tollervey, D.; Hurt, E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001, 21, 3405–3415. [Google Scholar] [CrossRef] [Green Version]
- Hedges, J.; Hedges, J.; West, M.; Johnson, A.W. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005, 24, 567–579. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.W.; Lund, E.; Dahlberg, J. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 2002, 27, 580–585. [Google Scholar] [CrossRef]
- Briones, E.; Briones, C.; Remacha, M.; Ballesta, J.P. The GTPase center protein L12 is required for correct ribosomal stalk assembly but not for Saccharomyces cerevisiae viability. J. Biol. Chem. 1998, 273, 31956–31961. [Google Scholar] [CrossRef] [Green Version]
- Tsay, Y.F.; Shankweiler, G.; Lake, J.; Woolford, J.L., Jr. Localization of Saccharomyces cerevisiae ribosomal protein L16 on the surface of 60 S ribosomal subunits by immunoelectron microscopy. J. Biol. Chem. 1994, 269, 7579–7586. [Google Scholar] [PubMed]
- Jakovljevic, J.; de Mayolo, P.A.; Miles, T.D.; Nguyen, T.M.; Léger-Silvestre, I.; Gas, N.; Woolford, J.L., Jr. The carboxy-terminal extension of yeast ribosomal protein S14 is necessary for maturation of 43S preribosomes. Mol. Cell. 2004, 14, 331–342. [Google Scholar] [CrossRef]
- Ford, C.L.; Randal-Whitis, L.; Ellis, S.R. Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res. 1999, 59, 704–710. [Google Scholar] [PubMed]
- Tabb-Massey, A.; Caffrey, J.M.; Logsden, P.; Taylor, S.; Trent, J.O.; Ellis, S.R. Ribosomal proteins Rps0 and Rps21 of Saccharomyces cerevisiae have overlapping functions in the maturation of the 3′ end of 18S rRNA. Nucleic Acids Res. 2003, 31, 6798–6805. [Google Scholar] [CrossRef] [Green Version]
- Van Beekvelt, C.A.; de Graaff-Vincent, M.; Faber, A.W.; van’t Riet, J.; Venema, J.; Raué, H.A. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 2001, 29, 5001–5008. [Google Scholar] [CrossRef]
- Leger-Silvestre, I.; Milkereit, P.; Ferreira-Cerca, S.; Saveanu, C.; Rousselle, J.C.; Choesmel, V.; Guinefoleau, C.; Gas, N.; Gleizes, P.E. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 2004, 23, 2336–2347. [Google Scholar] [CrossRef] [Green Version]
- Vallabhaneni, H.; Farabaugh, P.J. Accuracy modulating mutations of the ribosomal protein S4-S5 interface do not necessarily destabilize the rps4-rps5 protein-protein interaction. RNA 2009, 15, 1100–1109. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, D.; Gregory, S.T.; O’Connor, M. Error-prone and error-restrictive mutations affecting ribosomal protein S12. J. Mol. Biol. 2011, 410, 1–9. [Google Scholar] [CrossRef]
- Zaher, H.S.; Green, R. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol. Cell. 2010, 39, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Dresios, J.; Derkatch, I.L.; Liebman, S.W.; Synetos, D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000, 39, 7236–7244. [Google Scholar] [CrossRef]
- Deisenroth, C.; Franklin, D.A.; Zhang, Y. The evolution of the ribosomal protein-MDM2-p53 pathway. Cold Spring Harb. Perspect. Med. 2016, 6, a026138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Deisenroth, C.; Zhang, Y. RP-MDM2-p53 pathway: Linking ribosomal biogenesis and tumor surveillance. Trends Cancer 2016, 2, 191–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Xiong, X.; Sun, Y. The role of ribosomal proteins in the regulation of cell proliferation, tumorigenesis, and genomic integrity. Sci. China Life Sci. 2016, 59, 656–672. [Google Scholar] [CrossRef] [PubMed]
- Kampen, K.R.; Sulima, S.O.; Vereecke, S.; De Keersmaecker, K. Hallmarks of ribosomopathies. Nucleic Acids Res. 2020, 48, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- Arthurs, C.; Murtaza, B.N.; Thomson, C.; Dickens, K.; Henrique, R.; Patel, H.R.H.; Beltran, M.; Millar, M.; Thrasivoulou, C.; Ahmed, A. Expression of ribosomal proteins in normal and cancerous human prostate tissue. PLoS ONE 2017, 12, e0186047. [Google Scholar] [CrossRef] [Green Version]
- Nakhoul, H.; Ke, J.; Zhou, X.; Liao, W.; Zeng, S.X.; Lu, H. Ribosomopathies: Mechanisms of disease. Clin. Med. Insights. Blood Disord. 2014, 7, 7–16. [Google Scholar] [CrossRef]
- Narla, A.; Ebert, B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010, 115, 3196–3205. [Google Scholar] [CrossRef]
- De Keersmaecker, K. A novel mouse model provides insights into the neutropenia associated with the ribosomopathy Shwachman-Diamond syndrome. Haematologica 2015, 100, 1237–1239. [Google Scholar] [CrossRef] [Green Version]
- Vlachos, A.; Ball, S.; Dahl, N.; Alter, B.P.; Sheth, S.; Ramenghi, U.; Meerpohl, J.; Karlsson, S.; Liu, J.M.; Leblanc, T.; et al. Diagnosing and treating diamond blackfan anaemia: Results of an international clinical consensus conference. Br. J. Haematol. 2008, 142, 859–876. [Google Scholar] [CrossRef]
- Vlachos, A.; Muir, E. How I treat Diamond-Blackfan anemia. Blood 2010, 116, 3715–3723. [Google Scholar] [CrossRef] [Green Version]
- Horos, R.; von Lindern, M. Molecular mechanisms of pathology and treatment in Diamond Blackfan Anaemia. Br. J. Haematol. 2012, 159, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Padron, E.; Komrokji, R.; List, A.F. Biology and treatment of the 5q- syndrome. Expert Rev. Hematol. 2011, 4, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Finch, A.J.; Hilcenko, C.; Basse, N.; Drynan, L.F.; Goyenechea, B.; Menne, T.F.; González Fernández, A.; Simpson, P.; D’Santos, C.S.; Arends, M.J.; et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011, 25, 917–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, P.J.; Bessler, M. The genetics of dyskeratosis congenita. Cancer Genet. 2011, 204, 635–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiel, C.T.; Rauch, A. The molecular basis of the cartilage-hair hypoplasia-anauxetic dysplasia spectrum. Best. Pr. Res. Clin. Endocrinol. Metab. 2011, 25, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Schlump, J.U.; Stein, A.; Hehr, U.; Karen, T.; Möller-Hartmann, C.; Elcioglu, N.H.; Bogdanova, N.; Woike, H.F.; Lohmann, D.R.; Felderhoff-Mueser, U.; et al. Treacher Collins syndrome: Clinical implications for the paediatrician—a new mutation in a severely affected newborn and comparison with three further patients with the same mutation, and review of the literature. Eur. J. Pediatr. 2012, 171, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Sondalle, S.B.; Baserga, S.J. Human diseases of the SSU processome. Biochim. Biophys. Acta. 2014, 1842, 758–764. [Google Scholar] [CrossRef] [Green Version]
- Armistead, J.; Khatkar, S.; Meyer, B.; Mark, B.L.; Patel, N.; Coghlan, G.; Lamont, R.E.; Liu, S.; Wiechert, J.; Cattini, P.A.; et al. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am. J. Hum. Genet. 2009, 84, 728–739. [Google Scholar] [CrossRef] [Green Version]
- Meyer, B.; Wurm, J.P.; Kötter, P.; Leisegang, M.S.; Schilling, V.; Buchhaupt, M.; Held, M.; Bahr, U.; Karas, M.; Heckel, A.; et al. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Psi1191 in yeast 18S rRNA. Nucleic Acids Res. 2011, 39, 1526–1537. [Google Scholar] [CrossRef]
- Chagnon, P.; Michaud, J.; Mitchell, G.; Mercier, J.; Marion, J.F.; Drouin, E.; Rasquin-Weber, A.; Hudson, T.J.; Richter, A. A missense mutation (R565W) in cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am. J. Hum. Genet. 2002, 71, 1443–1449. [Google Scholar] [CrossRef] [Green Version]
- Freed, E.F.; Prieto, J.L.; McCann, K.L.; McStay, B.; Baserga, S.J. NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet. 2012, 8, e1002892. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhang, W.; Gao, J.; Chen, H.; Jiang, L.; Liu, D.; Cao, Y.; Zhao, S.; Qiu, Z.; Zeng, J. Downregulation of ribosomal protein S6 inhibits the growth of non-small cell lung cancer by inducing cell cycle arrest, rather than apoptosis. Cancer Lett. 2014, 354, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Ning, Q.; Pang, Y.; Shao, S.; Luo, M.; Zhao, L.; Hu, T.; Zhao, X. MicroRNA-147b suppresses the proliferation and invasion of non-small-cell lung cancer cells through downregulation of Wnt/beta-catenin signalling via targeting of RPS15A. Clin. Exp. Pharm. Physiol. 2020, 47, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Tsofack, S.P.; Meunier, L.; Sanchez, L.; Madore, J.; Provencher, D.; Mes-Masson, A.M.; Lebel, M. Low expression of the X-linked ribosomal protein S4 in human serous epithelial ovarian cancer is associated with a poor prognosis. BMC Cancer 2013, 13, 303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paquet, É.R.; Hovington, H.; Brisson, H.; Lacombe, C.; Larue, H.; Têtu, B.; Lacombe, L.; Fradet, Y.; Lebel, M. Low level of the X-linked ribosomal protein S4 in human urothelial carcinomas is associated with a poor prognosis. Biomark. Med. 2015, 9, 187–197. [Google Scholar] [CrossRef]
- Maruyama, Y.; Miyazaki, T.; Ikeda, K.; Okumura, T.; Sato, W.; Horie-Inoue, K.; Okamoto, K.; Takeda, S.; Inoue, S. Short hairpin RNA library-based functional screening identified ribosomal protein L31 that modulates prostate cancer cell growth via p53 pathway. PLoS ONE 2014, 9, e108743. [Google Scholar] [CrossRef]
- Fan, H.; Li, J.; Jia, Y.; Wu, J.; Yuan, L.; Li, M.; Wei, J.; Xu, B. Silencing of ribosomal protein L34 (RPL34) inhibits the proliferation and invasion of esophageal cancer cells. Oncol. Res. 2017, 25, 1061–1068. [Google Scholar] [CrossRef]
- Rao, S.; Cai, K.Q.; Stadanlick, J.E.; Greenberg-Kushnir, N.; Solanki-Patel, N.; Lee, S.Y.; Fahl, S.P.; Testa, J.R.; Wiest, D.L. Ribosomal protein Rpl22 controls the dissemination of T-cell lymphoma. Cancer Res. 2016, 76, 3387–3396. [Google Scholar] [CrossRef] [Green Version]
- Nieminen, T.T.; O’Donohue, M.F.; Wu, Y.; Lohi, H.; Scherer, S.W.; Paterson, A.D.; Ellonen, P.; Abdel-Rahman, W.M.; Valo, S.; Mecklin, J.P.; et al. Germline mutation of RPS20, encoding a ribosomal protein, causes predisposition to hereditary nonpolyposis colorectal carcinoma without DNA mismatch repair deficiency. Gastroenterology 2014, 147, 595–598. [Google Scholar] [CrossRef] [Green Version]
- Ruggero, D.; Shimamura, A. Marrow failure: A window into ribosome biology. Blood 2014, 124, 2784. [Google Scholar] [CrossRef] [Green Version]
- Burwick, N.; Shimamura, A.; Liu, J.M. Non-Diamond Blackfan anemia disorders of ribosome function: Shwachman Diamond syndrome and 5q- syndrome. Semin. Hematol. 2011, 48, 136–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dror, Y.; Donadieu, J.; Koglmeier, J.; Dodge, J.; Toiviainen-Salo, S.; Makitie, O.; Kerr, E.; Zeidler, C.; Shimamura, A.; Shah, N.; et al. Draft consensus guidelines for diagnosis and treatment of Shwachman-Diamond syndrome. Ann. N. Y. Acad. Sci. 2011, 1242, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Myers, K.C.; Davies, S.M.; Shimamura, A. Clinical and molecular pathophysiology of Shwachman-Diamond syndrome: An update. Hematol. Oncol. Clin. North Am. 2013, 27, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Ajore, R.; Raiser, D.; McConkey, M.; Jöud, M.; Boidol, B.; Mar, B.; Saksena, G.; Weinstock, D.M.; Armstrong, S.; Ellis, S.R.; et al. Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol. Med. 2017, 9, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Amsterdam, A.; Sadler, K.C.; Lai, K.; Farrington, S.; Bronson, R.T.; Lees, J.A.; Hopkins, N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004, 2, e139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazerounian, S.; Ciarlini, P.; Ghazvinian, R.; Alberich-Jorda, M.; Yuan, D.; Joshi, M.; Zhang, H.; Beggs, A.; Gazda, H.T. Increased tumorigenesis in ribosomal proteins L5 and S24 heterozygous mice. Blood 2013, 122, 1227. [Google Scholar] [CrossRef]
- Loreni, F.; Mancino, M.; Biffo, S. Translation factors and ribosomal proteins control tumor onset and progression: How? Oncogene 2014, 33, 2145–2156. [Google Scholar] [CrossRef] [Green Version]
- Henry, J.L.; Coggin, D.L.; King, C.R. High-level expression of the ribosomal protein L19 in human breast tumors that overexpress erbB-2. Cancer Res. 1993, 53, 1403–1408. [Google Scholar]
- Dressman, M.A.; Baras, A.; Malinowski, R.; Alvis, L.B.; Kwon, I.; Walz, T.M.; Polymeropoulos, M.H. Gene expression profiling detects gene amplification and differentiates tumor types in breast cancer. Cancer Res. 2003, 63, 2194–2199. [Google Scholar]
- Shi, Y.; Zhai, H.; Wang, X.; Han, Z.; Liu, C.; Lan, M.; Du, J.; Guo, C.; Zhang, Y.; Wu, K.; et al. Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp. Cell. Res. 2004, 296, 337–346. [Google Scholar] [CrossRef]
- Xie, X.; Guo, P.; Yu, H.; Wang, Y.; Chen, G. Ribosomal proteins: Insight into molecular roles and functions in hepatocellular carcinoma. Oncogene 2018, 37, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.D.; Xu, J. Ribosomal proteins and colorectal cancer. Curr. Genom. 2007, 8, 43–49. [Google Scholar]
- Kasai, H.; Nadano, D.; Hidaka, E.; Higuchi, K.; Kawakubo, M.; Sato, T.A.; Nakayama, J. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J. Histochem. Cytochem. 2003, 51, 567–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.J.; Chien, C.C.; Yang, S.H.; Chang, C.C.; Sun, H.L.; Cheng, Y.C.; Liu, C.C.; Lin, S.C.; Lin, C.M. Faecal ribosomal protein L19 is a genetic prognostic factor for survival in colorectal cancer. J. Cell. Mol. Med. 2008, 12, 1936–1943. [Google Scholar] [CrossRef] [PubMed]
- Bee, A.; Ke, Y.; Forootan, S.; Lin, K.; Beesley, C.; Forrest, S.E.; Foster, C.S. Ribosomal protein l19 is a prognostic marker for human prostate cancer. Clin. Cancer Res. 2006, 12, 2061–2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guimaraes, J.C.; Zavolan, M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016, 17, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hollis, A.R.; Starkey, M.P. MicroRNAs in equine veterinary science. Equine Vet. J. 2018, 50, 721–726. [Google Scholar] [CrossRef]
- Reza, A.M.M.T.; Choi, Y.J.; Han, S.G.; Song, H.; Park, C.; Hong, K.; Kim, J.H. Roles of microRNAs in mammalian reproduction: From the commitment of germ cells to peri-implantation embryos. Biol. Rev. 2019, 94, 415–438. [Google Scholar] [CrossRef]
- Rajman, M.; Schratt, G. MicroRNAs in neural development: From master regulators to fine-tuners. Development 2017, 144, 2310. [Google Scholar] [CrossRef] [Green Version]
- Burgos-Aceves, M.A.; Cohen, A.; Smith, Y.; Faggio, C. A potential microRNA regulation of immune-related genes in invertebrate haemocytes. Sci. Total Environ. 2018, 621, 302–307. [Google Scholar] [CrossRef]
- Nejad, C.; Stunden, H.J.; Gantier, M.P. A guide to miRNAs in inflammation and innate immune responses. FEBS J. 2018, 285, 3695–3716. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Lu, L.F. MicroRNA in immune Regulation. Curr. Top. Microbiol. Immunol. 2017, 410, 249–267. [Google Scholar] [PubMed]
- Kim, D.Y.; Sung, J.H. Regulatory role of microRNAs in the proliferation and differentiation of adipose-derived stem cells. Histol. Histopathol. 2017, 32, 1–10. [Google Scholar] [PubMed]
- Yu, X.; An, J.; Hua, Y.; Li, Z.; Yan, N.; Fan, W.; Su, C. MicroRNA-194 regulates keratinocyte proliferation and differentiation by targeting Grainyhead-like 2 in psoriasis. Pathol. Res. Pr. 2017, 213, 89–97. [Google Scholar] [CrossRef]
- Zhang, W.M.; Zhang, Z.R.; Yang, X.T.; Zhang, Y.G.; Gao, Y.S. Overexpression of miR21 promotes neural stem cell proliferation and neural differentiation via the Wnt/betacatenin signaling pathway in vitro. Mol. Med. Rep. 2018, 17, 330–335. [Google Scholar]
- Zhang, Y.; Shen, B.; Zhang, D.; Wang, Y.; Tang, Z.; Ni, N.; Jin, X.; Luo, M.; Sun, H.; Gu, P. miR-29a regulates the proliferation and differentiation of retinal progenitors by targeting Rbm8a. Oncotarget 2017, 8, 31993–32008. [Google Scholar] [CrossRef] [Green Version]
- Ran, X.; Xiao, C.-H.; Xiang, G.-M.; Ran, X.-Z. Regulation of embryonic stem cell self-renewal and differentiation by microRNAs. Cell. Reprogram. 2017, 19, 150–158. [Google Scholar] [CrossRef]
- Acunzo, M.; Croce, C.M. MicroRNA in cancer and cachexi—a mini-review. J. Infect. Dis. 2015, 212 (Suppl. 1), S74–S77. [Google Scholar] [CrossRef]
- Reza, A.M.M.T.; Choi, Y.J.; Yasuda, H.; Kim, J.H. Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci. Rep. 2016, 6, 38498. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, Z.; Shi, Y.; Fan, D. miRNAs in human cancers: The diagnostic and therapeutic implications. Curr. Pharm. Des. 2014, 20, 5336–5347. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.H.; Lin, W.C.; Tsai, K.W. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev. Mol. Med. 2014, 16, e1. [Google Scholar] [CrossRef] [PubMed]
- Teoh, S.L.; Das, S. The role of microRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr. Pharm. Des. 2017, 23, 1845–1859. [Google Scholar] [CrossRef] [PubMed]
- Deb, B.; Uddin, A.; Chakraborty, S. miRNAs and ovarian cancer: An overview. J. Cell. Physiol. 2018, 233, 3846–3854. [Google Scholar] [CrossRef]
- Qadir, M.I.; Faheem, A. miRNA: A diagnostic and therapeutic tool for pancreatic cancer. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 197–204. [Google Scholar] [CrossRef]
- Williams, M.; Cheng, Y.Y.; Blenkiron, C.; Reid, G. Exploring mechanisms of microRNA downregulation in cancer. MicroRNA 2017, 6, 2–16. [Google Scholar] [CrossRef]
- Quinlan, S.; Kenny, A.; Medina, M.; Engel, T.; Jimenez-Mateos, E.M. MicroRNAs in neurodegenerative diseases. Int. Rev. Cell. Mol. Biol. 2017, 334, 309–343. [Google Scholar]
- Molasy, M.; Walczak, A.; Szaflik, J.; Szaflik, J.P.; Majsterek, I. MicroRNAs in glaucoma and neurodegenerative diseases. J. Hum. Genet. 2017, 62, 105–112. [Google Scholar] [CrossRef]
- Rinchetti, P.; Rizzuti, M.; Faravelli, I.; Corti, S. MicroRNA metabolism and dysregulation in amyotrophic lateral sclerosis. Mol. Neurobiol. 2018, 55, 2617–2630. [Google Scholar] [CrossRef] [Green Version]
- Jiang, B.; Huo, Y.; Gu, Y.; Wang, J. The role of microRNAs in myopia. Graefes Arch. Clin. Exp. Ophthalmol. 2017, 255, 7–13. [Google Scholar] [CrossRef]
- Pan, Y.B.; Sun, Z.L.; Feng, D.F. The role of microRNA in traumatic brain injury. Neuroscience 2017, 367, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Baradan, R.; Hollander, J.M.; Das, S. Mitochondrial miRNAs in diabetes: Just the tip of the iceberg. Can. J. Physiol. Pharm. 2017, 95, 1156–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, L.C.; Huang, A.F.; Jia, H.; Liu, Y.; Xu, W.D. Role of microRNA-155 in rheumatoid arthritis. Int. J. Rheum. Dis. 2017, 20, 1631–1637. [Google Scholar] [CrossRef] [PubMed]
- Tavasolian, F.; Abdollahi, E.; Rezaei, R.; Momtazi-Borojeni, A.A.; Henrotin, Y.; Sahebkar, A. Altered expression of microRNAs in rheumatoid arthritis. J. Cell. Biochem. 2018, 119, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Pracht, K.; Mashreghi, M.F.; Jäck, H.M.; Radbruch, A.; Seliger, B. The role of the miR-148/-152 family in physiology and disease. Eur. J. Immunol. 2017, 47, 2026–2038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alipoor, S.D.; Adcock, I.M.; Garssen, J.; Mortaz, E.; Varahram, M.; Mirsaeidi, M.; Velayati, A. The roles of miRNAs as potential biomarkers in lung diseases. Eur. J. Pharm. 2016, 791, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, C.; Balmayor, E.R.; van Griensven, M. miRNAs related to skeletal diseases. Stem. Cells. Dev. 2016, 25, 1261–1281. [Google Scholar] [CrossRef]
- Jung, H.J.; Suh, Y. Circulating miRNAs in ageing and ageing-related diseases. J. Genet. Genom. 2014, 41, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Vijayan, M.; Bhatti, J.S.; Reddy, P.H. MicroRNAs as peripheral biomarkers in aging and age-related diseases. Prog. Mol. Biol. Transl. Sci. 2017, 146, 47–94. [Google Scholar]
- Cheng, H.S.; Njock, M.S.; Khyzha, N.; Dang, L.T.; Fish, J.E. Noncoding RNAs regulate NF-κB signaling to modulate blood vessel inflammation. Front. Genet. 2014, 5, 422. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Meng, X.; Li, G.; Zhou, Q.; Xiao, J. Circular RNAs in cardiovascular diseases. Adv. Exp. Med. Biol. 2018, 1087, 191–204. [Google Scholar] [PubMed]
- Bandara, K.V.; Michael, M.Z.; Gleadle, J.M. MicroRNA biogenesis in hypoxia. MicroRNA 2017, 6, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Harden, J.T.; Krams, S.M. Micro-RNAs in transplant tolerance. Curr. Opin. Organ Transpl. 2018, 23, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, G.; Liu, M.-L. Microvesicles as emerging biomarkers and therapeutic targets in cardiometabolic diseases. Genom. Proteom. Bioinf. 2018, 16, 50–62. [Google Scholar] [CrossRef]
- Arnold, M.L.; Melentijevic, I.; Smart, A.J.; Driscoll, M. Q&A: Trash talk: Disposal and remote degradation of neuronal garbage. BMC Biol. 2018, 16, 17. [Google Scholar]
- Qu, Y.; Liu, H.; Lv, X.; Liu, Y.; Wang, X.; Zhang, M.; Zhang, X.; Li, Y.; Lou, Q.; Li, S. MicroRNA-16-5p overexpression suppresses proliferation and invasion as well as triggers apoptosis by targeting VEGFA expression in breast carcinoma. Oncotarget 2017, 8, 72400–72410. [Google Scholar] [CrossRef]
- Ruan, L.; Qian, X. MiR-16-5p inhibits breast cancer by reducing AKT3 to restrain NF-κB pathway. Biosci. Rep. 2019, 39 BSR20191611, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Cheng, B.; Ding, F.; Huang, C.Y.; Xiao, H.; Fei, F.Y.; Li, J. Role of miR-16-5p in the proliferation and metastasis of hepatocellular carcinoma. Eur. Rev. Med. Pharm. Sci. 2019, 23, 137–145. [Google Scholar]
- Munson, P.B.; Hall, E.M.; Farina, N.H.; Pass, H.I.; Shukla, A. Exosomal miR-16-5p as a target for malignant mesothelioma. Sci. Rep. 2019, 9, 11688. [Google Scholar] [CrossRef] [Green Version]
- Krell, A.; Wolter, M.; Stojcheva, N.; Hertler, C.; Liesenberg, F.; Zapatka, M.; Weller, M.; Malzkorn, B.; Reifenberger, G. MiR-16-5p is frequently down-regulated in astrocytic gliomas and modulates glioma cell proliferation, apoptosis and response to cytotoxic therapy. Neuropathol. Appl. Neurobiol. 2019, 45, 441–458. [Google Scholar] [CrossRef]
- Chava, S.; Reynolds, C.P.; Pathania, A.S.; Gorantla, S.; Poluektova, L.Y.; Coulter, D.W.; Gupta, S.C.; Pandey, M.K.; Challagundla, K.B. miR-15a-5p, miR-15b-5p, and miR-16-5p inhibit tumor progression by directly targeting MYCN in neuroblastoma. Mol. Oncol. 2019, 14, 180–196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, K.; Ren, T.; Huang, Y.; Tang, X.; Guo, W. miR-16-5p inhibits chordoma cell proliferation, invasion and metastasis by targeting Smad3. Cell Death Dis. 2018, 9, 680. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jia, J.; Liu, X.; Yang, S.; Ye, S.; Yang, W.; Zhang, Y. MicroRNA-16-5p controls development of osteoarthritis by targeting SMAD3 in chondrocytes. Curr. Pharm. Des. 2015, 21, 5160–5167. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Huang, Q.; Zhu, H. Melatonin inhibits the proliferation of gastric cancer cells through regulating the miR-16-5p-Smad3 pathway. DNA Cell. Biol. 2018, 37, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Zhang, Z.; Qin, S.; Li, C.; Dong, Y. MicroRNA-16-5p inhibits osteoclastogenesis in giant cell tumor of bone. Biomed. Res. Int. 2017, 2017, 3173547. [Google Scholar] [CrossRef] [Green Version]
- Dunaeva, M.; Blom, J.; Thurlings, R.; Pruijn, G.J.M. Circulating serum miR-223-3p and miR-16-5p as possible biomarkers of early rheumatoid arthritis. Clin. Exp. Immunol. 2018, 193, 376–385. [Google Scholar] [CrossRef] [Green Version]
- Venza, M.; Visalli, M.; Beninati, C.; Benfatto, S.; Teti, D.; Venza, I. miR-92a-3p and MYCBP2 are involved in MS-275-induced and c-myc-mediated TRAIL-sensitivity in melanoma cells. Int. Immunopharmacol. 2016, 40, 235–243. [Google Scholar] [CrossRef]
- Casadei, L.; Calore, F.; Creighton, C.J.; Guescini, M.; Batte, K.; Iwenofu, O.H.; Zewdu, A.; Braggio, D.A.; Bill, K.L.; Fadda, P.; et al. Exosome-derived miR-25-3p and miR-92a-3p stimulate liposarcoma progression. Cancer Res. 2017, 77, 3846–3856. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Zhang, Y.; Liu, N.; Zhao, S.; Kong, Y.; Yuan, L. miR-92a-3p exerts various effects in glioma and glioma stem-like cells specifically targeting CDH1/beta-catenin and Notch-1/Akt signaling pathways. Int. J. Mol. Sci. 2016, 17, 1799. [Google Scholar] [CrossRef]
- Sharifi, M.; Salehi, R. Blockage of miR-92a-3p with locked nucleic acid induces apoptosis and prevents cell proliferation in human acute megakaryoblastic leukemia. Cancer Gene 2016, 23, 29–35. [Google Scholar] [CrossRef]
- Ahmadi, S.; Sharifi, M.; Salehi, R. Locked nucleic acid inhibits miR-92a-3p in human colorectal cancer, induces apoptosis and inhibits cell proliferation. Cancer Gene 2016, 23, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.; Jiang, W.; Zhou, L.; Chen, Z. Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic stage and grade of colorectal cancer. Transl. Oncol. 2018, 11, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Zhang, Z.; Hu, S.; Zhang, Z.; Chang, Z.; Huang, Z.; Liao, W.; Kang, Y. Exosomes derived from miR-92a-3p-overexpressing human mesenchymal stem cells enhance chondrogenesis and suppress cartilage degradation via targeting WNT5A. Stem Cell Res. 2018, 9, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rong, X.; Jia, L.; Hong, L.; Pan, L.; Xue, X.; Zhang, C.; Lu, J.; Jin, Z.; Qiu, H.; Wu, R. Serum miR-92a-3p as a new potential biomarker for diagnosis of Kawasaki disease with coronary artery lesions. J. Cardiovasc. Transl. Res. 2017, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shang, S.; Wang, J.; Zhang, T.; Nie, F.; Song, X.; Zhao, H.; Zhu, C.; Zhang, R.; Hao, D. Identification of miR-22-3p, miR-92a-3p, and miR-137 in peripheral blood as biomarker for schizophrenia. Psychiatry Res. 2018, 265, 70–76. [Google Scholar] [CrossRef]
- Kim, B.S.; Jung, J.Y.; Jeon, J.Y.; Kim, H.A.; Suh, C.H. Circulating hsa-miR-30e-5p, hsa-miR-92a-3p, and hsa-miR-223-3p may be novel biomarkers in systemic lupus erythematosus. HLA 2016, 88, 187–193. [Google Scholar] [CrossRef]
- He, J.R.; Zhang, Y.; Lu, W.J.; Liang, H.B.; Tu, X.Q.; Ma, F.Y.; Yang, G.Y.; Zeng, L.L. Age-related frontal periventricular white matter hyperintensities and miR-92a-3p are associated with early-onset post-stroke depression. Front. Aging Neurosci. 2017, 9, 328. [Google Scholar] [CrossRef] [Green Version]
- Nabavi, N.; Saidy, N.R.N.; Venalainen, E.; Haegert, A.; Parolia, A.; Xue, H.; Wang, Y.; Wu, R.; Dong, X.; Collins, C.; et al. miR-100-5p inhibition induces apoptosis in dormant prostate cancer cells and prevents the emergence of castration-resistant prostate cancer. Sci. Rep. 2017, 7, 4079. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Lin, C.; Quan, J.; Lai, Y.; He, T.; Zhou, L.; Pan, X.; Wu, X.; Wang, Y.; Ni, L.; et al. Oncogenic miR-100-5p is associated with cellular viability, migration and apoptosis in renal cell carcinoma. Mol. Med. Rep. 2017, 16, 5023–5030. [Google Scholar] [CrossRef] [Green Version]
- Jakob, M.; Mattes, L.M.; Küffer, S.; Unger, K.; Hess, J.; Bertlich, M.; Haubner, F.; Ihler, F.; Canis, M.; Weiss, B.G.; et al. MicroRNA expression patterns in oral squamous cell carcinoma: Hsa-mir-99b-3p and hsa-mir-100-5p as novel prognostic markers for oral cancer. Head Neck 2019, 41, 3499–3515. [Google Scholar] [CrossRef]
- Lai, Y.; Kacal, M.; Kanony, M.; Stukan, I.; Jatta, K.; Kis, L.; Norberg, E.; Vakifahmetoglu-Norberg, H.; Lewensohn, R.; Hydbring, P.; et al. miR-100-5p confers resistance to ALK tyrosine kinase inhibitors Crizotinib and Lorlatinib in EML4-ALK positive NSCLC. Biochem. Biophys. Res. Commun. 2019, 511, 260–265. [Google Scholar] [CrossRef] [PubMed]
- He, Q.L.; Qin, S.Y.; Tao, L.; Ning, H.J.; Jiang, H.X. Prognostic value and prospective molecular mechanism of miR-100-5p in hepatocellular carcinoma: A comprehensive study based on 1,258 samples. Oncol. Lett. 2019, 18, 6126–6142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Luo, H.; Chen, Y.; Wu, Q.; Xiong, Y.; Zhu, J.; Diao, Y.; Wu, Z.; Miao, J.; Wan, J. MicroRNAs 99b-5p/100-5p regulated by endoplasmic reticulum stress are involved in abeta-induced pathologies. Front. Aging Neurosci. 2015, 7, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Li, C.; Xu, H.; Duan, Z.; Liu, Y.; Zeng, R.; Li, M.; Wang, B. AKT-dependent hyperproliferation of keratinocytes in familial hidradenitis suppurativa with a NCSTN mutation: A potential role of defective miR-100-5p. Br. J. Derm. 2019, 182, 500–502. [Google Scholar] [CrossRef]
- Wang, J.; Liu, L.; Sun, Y.; Xue, Y.; Qu, J.; Pan, S.; Li, H.; Qu, H.; Wang, J.; Zhang, J. miR-615-3p promotes proliferation and migration and inhibits apoptosis through its potential target CELF2 in gastric cancer. Biomed. Pharm. 2018, 101, 406–413. [Google Scholar] [CrossRef]
- Laursen, E.B.; Fredsøe, J.; Schmidt, L.; Strand, S.H.; Kristensen, H.; Rasmussen, A.K.I.; Daugaard, T.F.; Mouritzen, P.; Høyer, S.; Kristensen, G.; et al. Elevated miR-615-3p expression predicts adverse clinical outcome and promotes proliferation and migration of prostate cancer cells. Am. J. Pathol. 2019, 189, 2377–2388. [Google Scholar] [CrossRef] [Green Version]
- Pu, H.Y.; Xu, R.; Zhang, M.Y.; Yuan, L.J.; Hu, J.Y.; Huang, G.L.; Wang, H.Y. Identification of microRNA-615-3p as a novel tumor suppressor in non-small cell lung cancer. Oncol. Lett. 2017, 13, 2403–2410. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Jia, Y.; Jia, L.; Li, T.; Yang, L.; Zhang, G. MicroRNA 615-3p inhibits the tumor growth and metastasis of NSCLC via inhibiting IGF2. Oncol. Res. 2019, 27, 269–279. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Pan, S.; Yang, T.; Sun, X.; Wang, Y.; Shi, X.; Zhao, X.; Guo, J.; Zhang, X. LINC00657 played oncogenic roles in esophageal squamous cell carcinoma by targeting miR-615-3p and JunB. Biomed. Pharm. 2018, 108, 316–324. [Google Scholar] [CrossRef]
- Mukai, R.; Tomimaru, Y.; Nagano, H.; Eguchi, H.; Mimori, K.; Tomokuni, A.; Asaoka, T.; Wada, H.; Kawamoto, K.; Marubashi, S.; et al. miR-615-3p expression level in bone marrow is associated with tumor recurrence in hepatocellular carcinoma. Mol. Clin. Oncol. 2015, 3, 487–494. [Google Scholar] [CrossRef]
- Hu, Y.; Xie, H.; Liu, Y.; Liu, W.; Liu, M.; Tang, H. miR-484 suppresses proliferation and epithelial-mesenchymal transition by targeting ZEB1 and SMAD2 in cervical cancer cells. Cancer Cell. Int. 2017, 17, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Wu, F.; Liu, Y.; Zhao, Q.; Tang, H. DNMT1 recruited by EZH2-mediated silencing of miR-484 contributes to the malignancy of cervical cancer cells through MMP14 and HNF1A. Clin. Epigenetics 2019, 11, 186. [Google Scholar] [CrossRef] [PubMed]
- Zare, A.; Ahadi, A.; Larki, P.; Omrani, M.D.; Zali, M.R.; Alamdari, N.M.; Ghaedi, H. The clinical significance of miR-335, miR-124, miR-218 and miR-484 downregulation in gastric cancer. Mol. Biol. Rep. 2018, 45, 1587–1595. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Feng, J.; Yang, S.; Huang, X.; Liao, Y.; Hu, Z.; Luo, M. miR-484/MAP2/c-Myc-positive regulatory loop in glioma promotes tumor-initiating properties through ERK1/2 signaling. J. Mol. Histol. 2018, 49, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ding, Z.L.; Zheng, Y.L.; Wang, W. MiR-484 promotes non-small-cell lung cancer (NSCLC) progression through inhibiting Apaf-1 associated with the suppression of apoptosis. Biomed. Pharm. 2017, 96, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Long, B.; Jiao, J.Q.; Wang, J.X.; Liu, J.P.; Li, Q.; Li, P.F. miR-484 regulates mitochondrial network through targeting Fis1. Nat. Commun. 2012, 3, 781. [Google Scholar] [CrossRef]
- Lu, X.; Lu, J. The significance of detection of serum miR-423-5p and miR-484 for diagnosis of colorectal cancer. Clin. Lab. 2015, 61, 187–190. [Google Scholar] [CrossRef]
- Merhautova, J.; Hezova, R.; Poprach, A.; Kovarikova, A.; Radova, L.; Svoboda, M.; Vyzula, R.; Demlova, R.; Slaby, O. miR-155 and miR-484 are associated with time to progression in metastatic renal cell carcinoma treated with sunitinib. Biomed. Res. Int. 2015, 2015, 941980. [Google Scholar] [CrossRef] [Green Version]
- Ye, F.G.; Song, C.G.; Cao, Z.G.; Xia, C.; Chen, D.N.; Chen, L.; Li, S.; Qiao, F.; Ling, H.; Yao, L. Cytidine deaminase axis modulated by miR-484 differentially regulates cell proliferation and chemoresistance in breast cancer. Cancer Res. 2015, 75, 1504–1515. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Bao, H.; Zhang, S.; Li, R.; Chen, L.; Zhu, Y. miR-186-5p promotes apoptosis by targeting IGF-1 in SH-SY5Y OGD/R model. Int. J. Biol. Sci. 2018, 14, 1791–1799. [Google Scholar] [CrossRef]
- Silva, M.M.; Rodrigues, B.; Fernandes, J.; Santos, S.D.; Carreto, L.; Santos, M.A.S.; Pinheiro, P.; Carvalho, A.L. MicroRNA-186-5p controls GluA2 surface expression and synaptic scaling in hippocampal neurons. Proc. Natl. Acad. Sci. USA 2019, 116, 5727–5736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Wu, J.; Wei, W.; Cai, X.; Yan, J.; Song, J.; Wang, C.; Wang, J. Association of serum miR-186-5p with the prognosis of acute coronary syndrome patients after percutaneous coronary intervention. Front. Physiol. 2019, 10, 686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, D.X.; Xiao, Y.; Wang, C.J.; Jiang, H.; Gao, Y.; Yuan, B.; Zhang, J.B. Regulation of FSH expression by differentially expressed miR-186-5p in rat anterior adenohypophyseal cells. PLoS ONE 2018, 13, e0194300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Zhang, W.; Mao, J.; Xu, Z.; Fan, M. miR-186-5p functions as a tumor suppressor in human osteosarcoma by targeting FOXK1. Cell. Physiol. Biochem. 2019, 52, 553–564. [Google Scholar]
- Cao, Q.; Wang, Z.; Wang, Y.; Liu, F.; Dong, Y.; Zhang, W.; Wang, L.; Ke, Z. TBL1XR1 promotes migration and invasion in osteosarcoma cells and is negatively regulated by miR-186-5p. Am. J. Cancer Res. 2018, 8, 2481–2493. [Google Scholar]
- Li, J.; Xia, L.; Zhou, Z.; Zuo, Z.; Xu, C.; Song, H.; Cai, J. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch. Biochem. Biophys. 2018, 640, 53–60. [Google Scholar] [CrossRef]
- Zhu, K.; Su, Y.; Xu, B.; Wang, Z.; Sun, H.; Wang, L.; Sun, C.; He, X. MicroRNA-186-5p represses neuroblastoma cell growth via downregulation of Eg5. Am. J. Transl. Res. 2019, 11, 2245–2256. [Google Scholar]
- Feng, H.; Zhang, Z.; Qing, X.; French, S.W.; Liu, D. miR-186-5p promotes cell growth, migration and invasion of lung adenocarcinoma by targeting PTEN. Exp. Mol. Pathol. 2019, 108, 105–113. [Google Scholar] [CrossRef]
- Jones, D.Z.; Schmidt, M.L.; Suman, S.; Hobbing, K.R.; Barve, S.S.; Gobejishvili, L.; Brock, G.; Klinge, C.M.; Rai, S.N.; Park, J.; et al. Micro-RNA-186-5p inhibition attenuates proliferation, anchorage independent growth and invasion in metastatic prostate cancer cells. BMC Cancer 2018, 18, 421. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, X.; Sun, W.; Yue, S.; Yang, J.; Li, J.; Ma, B.; Wang, J.; Yang, X.; Pu, M.; et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017, 397, 33–42. [Google Scholar] [CrossRef]
- Lv, G.; Wu, M.; Wang, M.; Jiang, X.; Du, J.; Zhang, K.; Li, D.; Ma, N.; Peng, Y.; Wang, L.; et al. miR-320a regulates high mobility group box 1 expression and inhibits invasion and metastasis in hepatocellular carcinoma. Liver Int. 2017, 37, 1354–1364. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Sun, Q.; Yu, Z.; Mao, S.; Jin, Y.; Li, J.; Jiang, Z.; Zhang, Y.; Chen, M.; Chen, P.; et al. miR-320a-3p/ELF3 axis regulates cell metastasis and invasion in non-small cell lung cancer via PI3K/Akt pathway. Gene 2018, 670, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yu, L.; Liu, J.; Bian, X.; Shi, C.; Sun, C.; Zhou, X.; Wen, Y.; Hua, D.; Zhao, S.; et al. miR-320a functions as a suppressor for gliomas by targeting SND1 and beta-catenin, and predicts the prognosis of patients. Oncotarget 2017, 8, 19723–19737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zeng, J.; Pan, J.; Geng, X.; Li, L.; Wu, J.; Song, P.; Wang, Y.; Liu, J.; Wang, L. miR-320a inhibits gastric carcinoma by targeting activity in the FoxM1-P27KIP1 axis. Oncotarget 2016, 7, 29275–29786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Liu, H.; Shao, J.; Xing, G. miR-320a serves as a negative regulator in the progression of gastric cancer by targeting RAB14. Mol. Med. Rep. 2017, 16, 2652–2658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Q.; Hu, J.X.; Li, Y.J.; Xie, N.; Song, D.D.; Zhao, W.; Yan, Y.F.; Li, B.S.; Wang, P.Y.; Xie, S.Y. miR-320a effectively suppresses lung adenocarcinoma cell proliferation and metastasis by regulating STAT3 signals. Cancer Biol. 2017, 18, 142–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, N.; Wang, C.; Zhuang, Z.; Hou, J.; Liu, X.; Wu, Y.; Liu, H.; Huang, H. Decreased miR-320a promotes invasion and metastasis of tumor budding cells in tongue squamous cell carcinoma. Oncotarget 2016, 7, 65744–65757. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Wu, D.; Wang, J.; Li, Y.; Chai, X.; Kang, Q. miR-320a regulates cell proliferation and apoptosis in multiple myeloma by targeting pre-B-cell leukemia transcription factor 3. Biochem. Biophys. Res. Commun. 2016, 473, 1315–1320. [Google Scholar] [CrossRef]
- Yu, J.; Wang, L.; Yang, H.; Ding, D.; Zhang, L.; Wang, J.; Chen, Q.; Zou, Q.; Jin, Y.; Liu, X. Rab14 suppression mediated by miR-320a inhibits cell proliferation, migration and invasion in breast cancer. J. Cancer 2016, 7, 2317–2326. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Zhao, H.; Dong, T.; Zhou, H.; Wang, L.; Huang, A.; Feng, B.; Quan, Y.; Jin, R.; Zhang, W.; et al. miR-320a suppresses colorectal cancer progression by targeting Rac1. Carcinogenesis 2014, 35, 886–895. [Google Scholar] [CrossRef] [Green Version]
- Shang, C.; Zhang, H.; Guo, Y.; Hong, Y.; Liu, Y.; Xue, Y. miR-320a down-regulation mediates bladder carcinoma invasion by targeting ITGB3. Mol. Biol. Rep. 2014, 41, 2521–2527. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Wang, J.; Yin, Z.; Zhao, Y.; Hou, H.; Fan, J.; Li, H.; Wen, Z.; Tang, J.; Wang, Y.; et al. miR-320a induces diabetic nephropathy via inhibiting MafB. Aging 2019, 11, 3055–3079. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Chen, X.; Gao, Z.Y.; Liu, K.; Hou, Y.; Zheng, J. The role of miR-320a and IL-1beta in human chondrocyte degradation. Bone Jt. Res. 2017, 6, 196–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De-Ugarte, L.; Balcells, S.; Nogues, X.; Grinberg, D.; Diez-Perez, A.; Garcia-Giralt, N. Pro-osteoporotic miR-320a impairs osteoblast function and induces oxidative stress. PLoS ONE 2018, 13, e0208131. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Nie, Z.K.; Li, F.; Guo, H.M.; Yang, X.L.; Ding, S.F. miR-320a was highly expressed in postmenopausal osteoporosis and acts as a negative regulator in MC3T3E1 cells by reducing MAP9 and inhibiting PI3K/AKT signaling pathway. Exp. Mol. Pathol. 2019, 110, 104282. [Google Scholar] [CrossRef]
- Yin, Z.; Zhao, Y.; Li, H.; Yan, M.; Zhou, L.; Chen, C.; Wang, D.W. miR-320a mediates doxorubicin-induced cardiotoxicity by targeting VEGF signal pathway. Aging 2016, 8, 192–207. [Google Scholar] [CrossRef] [Green Version]
- Gao, T.; Deng, M.; Wang, Q. MiRNA-320a inhibits trophoblast cell invasion by targeting estrogen-related receptor-gamma. J. Obs. Gynaecol. Res. 2018, 44, 756–763. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Y.; Yang, S.; Li, H.; Zhao, G.; Wang, F.; Yang, L.; Wang, D.W. miR-320a contributes to atherogenesis by augmenting multiple risk factors and down-regulating SRF. J. Cell. Mol. Med. 2015, 19, 970–985. [Google Scholar] [CrossRef]
- Sommariva, E.; D’Alessandra, Y.; Farina, F.M.; Casella, M.; Cattaneo, F.; Catto, V.; Chiesa, M.; Stadiotti, I.; Brambilla, S.; Dello Russo, A.; et al. miR-320a as a potential novel circulating biomarker of arrhythmogenic cardiomyopathy. Sci. Rep. 2017, 7, 4802. [Google Scholar] [CrossRef]
- Zhang, C.L.; Wang, H.; Yan, C.Y.; Gao, X.F.; Ling, X.J. Deregulation of RUNX2 by miR-320a deficiency impairs steroidogenesis in cumulus granulosa cells from polycystic ovary syndrome (PCOS) patients. Biochem. Biophys. Res. Commun. 2017, 482, 1469–1476. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Q.; Xu, J.; Zhang, X.; Zhang, H.; Xiang, Y.; Fang, C.; Wang, T.; Xia, S.; Zhang, Q.; et al. The aberrantly expressed miR-193b-3p contributes to preeclampsia through regulating transforming growth factor-beta signaling. Sci. Rep. 2016, 6, 19910. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qin, J.; Su, Y. miR-193b-3p possesses anti-tumor activity in ovarian carcinoma cells by targeting p21-activated kinase 3. Biomed. Pharm. 2017, 96, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhuang, Q.; Hu, G.; Geng, S. MORC4 is a novel breast cancer oncogene regulated by miR-193b-3p. J. Cell. Biochem. 2019, 120, 4634–4643. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.R.; Yeh, H.C.; Wang, W.J.; Ke, H.L.; Lin, H.H.; Hsu, W.C.; Chao, S.Y.; Hour, T.C.; Wu, W.J.; Pu, Y.S.; et al. miR-193b mediates CEBPD-induced cisplatin sensitization through targeting ETS1 and cyclin D1 in human urothelial carcinoma cells. J. Cell. Biochem. 2017, 118, 1563–1573. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Dou, Q.; Ha, X. Let-7a-5p inhibits BMSCs osteogenesis in postmenopausal osteoporosis mice. Biochem. Biophys. Res. Commun. 2019, 510, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Li, J.P.; Liao, X.H.; Xiang, Y.; Yao, A.; Song, R.H.; Zhang, Z.J.; Huang, F.; Dai, Z.T.; Zhang, T.C. Hyperoside and let-7a-5p synergistically inhibits lung cancer cell proliferation via inducing G1/S phase arrest. Gene. 2018, 679, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Yu, S.; Yuan, T.; Yao, S.; Zhang, L. Exogenous let-7a-5p induces A549 lung cancer cell death through BCL2L1-mediated PI3Kgamma signaling pathway. Front. Oncol. 2019, 9, 808. [Google Scholar] [CrossRef]
- Wu, W.Y.; Tao, S.Q.; Wang, X.N.; Lobie, P.E.; Wu, Z.S. XIAP 3′-untranslated region serves as a competitor for HMGA2 by arresting endogenous let-7a-5p in human hepatocellular carcinoma. Tumour. Biol. 2017, 39, 1010428317719578. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.P.; Huang, C.C.; Yeh, K.T.; Ke, T.W.; Wei, P.L.; Yang, J.R.; Cheng, Y.W. Down-regulation of let-7a-5p predicts lymph node metastasis and prognosis in colorectal cancer: Implications for chemotherapy. Surg. Oncol. 2016, 25, 429–434. [Google Scholar] [CrossRef]
- Fasihi-Ramandi, M.; Moridnia, A.; Najafi, A.; Sharifi, M. Inducing apoptosis and decreasing cell proliferation in human acute promyelocytic leukemia through regulation expression of CASP3 by let-7a-5p blockage. Indian J. Hematol. Blood Transfus. 2018, 34, 70–77. [Google Scholar] [CrossRef]
- Wang, T.; Zhu, H.; Yang, S.; Fei, X. Let-7a-5p may participate in the pathogenesis of diabetic nephropathy through targeting HMGA2. Mol. Med. Rep. 2019, 19, 4229–4237. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Aizawa, N.; Enomoto, H.; Nishiguchi, S.; Toyoda, H.; Kumada, T.; Iio, E.; Ito, K.; Ogawa, S.; Isogawa, M.; et al. Circulating let-7 levels in serum correlate with the severity of hepatic fibrosis in chronic hepatitis C. Open Forum Infect. Dis. 2018, 5, ofy268. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Luo, H.; Li, X.; Tian, X.; Peng, B.; Liu, S.; Zhan, T.; Wan, Y.; Chen, W.; Li, Y.; et al. miR-331-3p functions as an oncogene by targeting ST7L in pancreatic cancer. Carcinogenesis 2018, 39, 1006–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, W.; Zhong, N.; Wang, L.; Yu, J.; Yin, F.; Zhang, K. miR-331-3p inhibition of the hepatocellular carcinoma (HCC) Bel-7402 cell line by down-regulation of E2F1. J. Nanosci. Nanotechnol. 2019, 19, 5476–5482. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, J.; Wang, D.; Peng, H.; Tan, X.; Xiong, D.; Huang, A.; Tang, H. Upregulated in hepatitis B virus-associated hepatocellular carcinoma cells, miR-331-3p promotes proliferation of hepatocellular carcinoma cells by targeting ING5. Oncotarget 2015, 6, 38093–38106. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Li, J.; Jin, B.; Wang, T.; Gu, J. Evaluation of miR-331-3p and miR-23b-3p as serum biomarkers for hepatitis c virus-related hepatocellular carcinoma at early stage. Clin. Res. Hepatol. Gastroenterol. 2019, 44, 21–28. [Google Scholar] [CrossRef]
- Gu, J.; Zhang, J.; Zheng, L.; Ajani, J.A.; Wu, X.; Ye, Y. Serum miR-331-3p predicts tumor recurrence in esophageal adenocarcinoma. Sci. Rep. 2018, 8, 14006. [Google Scholar] [CrossRef]
- Epis, M.R.; Giles, K.M.; Beveridge, D.J.; Richardson, K.L.; Candy, P.A.; Stuart, L.M.; Bentel, J.; Cohen, R.J.; Leedman, P.J. miR-331-3p and aurora kinase inhibitor II co-treatment suppresses prostate cancer tumorigenesis and progression. Oncotarget 2017, 8, 55116–55134. [Google Scholar] [CrossRef] [Green Version]
- Epis, M.R.; Giles, K.M.; Barker, A.; Kendrick, T.S.; Leedman, P.J. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J. Biol. Chem. 2009, 284, 24696–24704. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Sui, Y.; Zheng, X. miR-331-3p inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt and ERK1/2 pathways in colorectal cancer. Oncol. Rep. 2016, 35, 1075–1082. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhu, J.; Liu, Y.; Duan, C.; Chang, R.; Zhang, C. MicroRNA-331-3p inhibits epithelial-mesenchymal transition by targeting ErbB2 and VAV2 through the Rac1/PAK1/beta-catenin axis in non-small-cell lung cancer. Cancer Sci. 2019, 110, 1883–1896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epis, M.R.; Giles, K.M.; Candy, P.A.; Webster, R.J.; Leedman, P.J. miR-331-3p regulates expression of neuropilin-2 in glioblastoma. J. Neurooncol. 2014, 116, 67–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, T.; Shimada, K.; Asano, A.; Tatsumi, Y.; Yamaguchi, N.; Yamazaki, M.; Konishi, N. MicroRNA-331-3p suppresses cervical cancer cell proliferation and E6/E7 expression by targeting NRP2. Int. J. Mol. Sci. 2016, 17, 1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buranjiang, G.; Kuerban, R.; Abuduwanke, A.; Li, X.; Kuerban, G. MicroRNA-331-3p inhibits proliferation and metastasis of ovarian cancer by targeting RCC2. Arch. Med. Sci. 2019, 15, 1520–1529. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Fujii, T.; Itami, H.; Uchiyama, T.; Nakai, T.; Hatakeyama, K.; Sugimoto, A.; Miyake, M.; Nakai, Y.; Tanaka, N.; et al. NACC1, as a target of microRNA-331-3p, regulates cell proliferation in urothelial carcinoma cells. Cancers 2018, 10, 347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Guo, L.; Ji, J.; Zhang, J.; Zhang, J.; Chen, X.; Cai, Q.; Li, J.; Gu, Q.; Liu, B.; et al. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem. Biophys. Res. Commun. 2010, 398, 1–6. [Google Scholar] [CrossRef]
- Fan, W.; Liu, Y.; Li, C.; Qu, X.; Zheng, G.; Zhang, Q.; Pan, Z.; Wang, Y.; Rong, J. microRNA-331-3p maintains the contractile type of vascular smooth muscle cells by regulating TNF-alpha and CD14 in intracranial aneurysm. Neuropharmacology 2019, 164, 107858. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Z.; Jiang, C.; Jiang, X.; Zhang, J. MiR-92b-3p promotes neurite growth and functional recovery via the PTEN/AKT pathway in acute spinal cord injury. J. Cell. Physiol. 2019, 234, 23043–23052. [Google Scholar] [CrossRef]
- Hu, Z.Q.; Luo, J.F.; Yu, X.J.; Zhu, J.N.; Huang, L.; Yang, J.; Fu, Y.H.; Li, T.; Xue, Y.M.; Feng, Y.Q.; et al. Targeting myocyte-specific enhancer factor 2D contributes to the suppression of cardiac hypertrophic growth by miR-92b-3p in mice. Oncotarget 2017, 8, 92079–92089. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.J.; Huang, Y.Q.; Shan, Z.X.; Zhu, J.N.; Hu, Z.Q.; Huang, L.; Feng, Y.Q.; Geng, Q.S. MicroRNA-92b-3p suppresses angiotensin II-induced cardiomyocyte hypertrophy via targeting HAND2. Life Sci. 2019, 232, 116635. [Google Scholar] [CrossRef]
- Long, M.; Zhan, M.; Xu, S.; Yang, R.; Chen, W.; Zhang, S.; Shi, Y.; He, Q.; Mohan, M.; Liu, Q.; et al. miR-92b-3p acts as a tumor suppressor by targeting Gabra3 in pancreatic cancer. Mol. Cancer 2017, 16, 167. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.; Ren, M.; Lv, Z.; Yang, Y.; Wang, Z. miR-92b-3p promotes colorectal carcinoma cell proliferation, invasion, and migration by inhibiting FBXW7 in vitro and in vivo. DNA Cell. Biol. 2018, 37, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Fu, S.; Lin, X.; Zheng, J.; Pu, J.; Gu, Y.; Deng, W.; Liu, Y.; He, Z.; Liang, W.; et al. miR-92b-3p functions as a key gene in esophageal squamous cell cancer as determined by co-expression analysis. Onco. Targets 2019, 12, 8339–8353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Huo, B.; Wang, Y.; Cheng, C. Downregulation of microRNA-92b-3p suppresses proliferation, migration, and invasion of gastric cancer SGC-7901 cells by targeting Homeobox D10. J. Cell. Biochem. 2019, 120, 17405–17412. [Google Scholar] [CrossRef] [PubMed]
- Uotani, K.; Fujiwara, T.; Yoshida, A.; Iwata, S.; Morita, T.; Kiyono, M.; Yokoo, S.; Kunisada, T.; Takeda, K.; Hasei, J.; et al. Circulating microRNA-92b-3p as a novel biomarker for monitoring of synovial sarcoma. Sci. Rep. 2017, 7, 14634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Q.; Lu, X.; Huang, P.; Gao, C.; Zhao, X.; Xing, T.; Li, G.; Bao, S.; Zheng, H. Expression of miR-652-3p and effect on apoptosis and drug sensitivity in pediatric acute lymphoblastic leukemia. Biomed. Res. Int. 2018, 2018, 5724686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Q.L.; Zhan, D.M.; Chong, Y.K.; Ding, L.; Yang, Y.G. miR-652-3p promotes bladder cancer migration and invasion by targeting KCNN3. Eur. Rev. Med. Pharm. Sci. 2019, 23, 8806–8812. [Google Scholar]
- Yang, W.; Zhou, C.; Luo, M.; Shi, X.; Li, Y.; Sun, Z.; Zhou, F.; Chen, Z.; He, J. miR-652-3p is upregulated in non-small cell lung cancer and promotes proliferation and metastasis by directly targeting Lgl1. Oncotarget 2016, 7, 16703–16715. [Google Scholar] [CrossRef] [Green Version]
- Nam, R.K.; Benatar, T.; Amemiya, Y.; Wallis, C.J.D.; Romero, J.M.; Tsagaris, M.; Sherman, C.; Sugar, L.; Seth, A. MicroRNA-652 induces NED in LNCaP and EMT in PC3 prostate cancer cells. Oncotarget 2018, 9, 19159–19176. [Google Scholar] [CrossRef] [Green Version]
- Huang, R.; Hu, Z.; Cao, Y.; Li, H.; Zhang, H.; Su, W.; Xu, Y.; Liang, L.; Melgiri, N.D.; Jiang, L. miR-652-3p inhibition enhances endothelial repair and reduces atherosclerosis by promoting Cyclin D2 expression. EBioMedicine 2019, 40, 685–694. [Google Scholar] [CrossRef] [Green Version]
- You, Y.; Que, K.; Zhou, Y.; Zhang, Z.; Zhao, X.; Gong, J.; Liu, Z. MicroRNA-766-3p inhibits tumour progression by targeting Wnt3a in hepatocellular carcinoma. Mol. Cells 2018, 41, 830–841. [Google Scholar] [PubMed]
- Chen, C.; Xue, S.; Zhang, J.; Chen, W.; Gong, D.; Zheng, J.; Ma, J.; Xue, W.; Chen, Y.; Zhai, W.; et al. DNA-methylation-mediated repression of miR-766-3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma. Int. J. Cancer 2017, 141, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Kleemann, M.; Schneider, H.; Unger, K.; Sander, P.; Schneider, E.M.; Fischer-Posovszky, P.; Handrick, R.; Otte, K. miR-744-5p inducing cell death by directly targeting HNRNPC and NFIX in ovarian cancer cells. Sci. Rep. 2018, 8, 9020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Shi, F.; Zhang, W.; Zhou, Y.; Huang, J. miR-744-5p inhibits non-small cell lung cancer proliferation and invasion by directly targeting PAX2. Technol. Cancer Res. Treat. 2019, 18, 1533033819876913. [Google Scholar] [CrossRef] [Green Version]
- Cai, B.; Ma, M.; Chen, B.; Li, Z.; Abdalla, B.A.; Nie, Q.; Zhang, X. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death Dis. 2018, 9, 367. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.P.; Wang, W.W.; Lu, W.Y.; Shang, A.Q. The mechanism of miR-16-5p protection on LPS-induced A549 cell injury by targeting CXCR3. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1200–1206. [Google Scholar] [CrossRef] [Green Version]
- Cun, J.; Yang, Q. Bioinformatics-based interaction analysis of miR-92a-3p and key genes in tamoxifen-resistant breast cancer cells. Biomed. Pharm. 2018, 107, 117–128. [Google Scholar] [CrossRef]
- Yin, J.; Zhuang, G.; Zhu, Y.; Hu, X.; Zhao, H.; Zhang, R.; Guo, H.; Fan, X.; Cao, Y. MiR-615-3p inhibits the osteogenic differentiation of human lumbar ligamentum flavum cells via suppression of osteogenic regulators GDF5 and FOXO1. Cell. Biol. Int. 2017, 41, 779–786. [Google Scholar] [CrossRef]
- Yan, T.; Ooi, W.F.; Qamra, A.; Cheung, A.; Ma, D.; Sundaram, G.M.; Xu, C.; Xing, M.; Poon, L.; Wang, J.; et al. HoxC5 and miR-615-3p target newly evolved genomic regions to repress hTERT and inhibit tumorigenesis. Nat. Commun. 2018, 9, 100. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Mauer, A.S.; Kumar, S.; Mott, J.L.; Malhi, H. Mmu-miR-615-3p regulates lipoapoptosis by inhibiting C/EBP homologous protein. PLoS ONE 2014, 9, e109637. [Google Scholar] [CrossRef]
- Zhuang, Z.; Sun, C.; Gong, H. High serum miR-484 expression is associated with the diagnosis and prognosis of patients with non-small cell lung cancer. Exp. Med. 2019, 18, 4095–4102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujitani, M.; Fujitani, M.; Zhang, S.; Fujiki, R.; Fujihara, Y.; Yamashita, T. A chromosome 16p13.11 microduplication causes hyperactivity through dysregulation of miR-484/protocadherin-19 signaling. Mol. Psychiatry 2017, 22, 364–374. [Google Scholar] [PubMed]
- Liu, H.; Li, S.; Jiang, W.; Li, Y. miR-484 protects rat myocardial cells from ischemia-reperfusion injury by inhibiting caspase-3 and caspase-9 during apoptosis. Korean Circ. J. 2019, 50, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Mo, H.; Liu, C.; Wu, B.; Wu, Z.; Li, X.; Li, T.; He, S.; Li, S.; You, Q. Inhibition of miR-186-5p contributes to high glucose-induced injury in AC16 cardiomyocytes. Exp. Med. 2018, 15, 627–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zheng, W.; Pan, Y.; Hu, J. Low expression of miR-186-5p regulates cell apoptosis by targeting toll-like receptor 3 in high glucose-induced cardiomyocytes. J. Cell. Biochem. 2019, 120, 9532–9538. [Google Scholar] [CrossRef]
- Meng, F.; Li, Z.; Zhang, Z.; Yang, Z.; Kang, Y.; Zhao, X.; Long, D.; Hu, S.; Gu, M.; He, S.; et al. microRNA-193b-3p regulates chondrogenesis and chondrocyte metabolism by targeting HDAC3. Theranostics 2018, 8, 2862–2883. [Google Scholar] [CrossRef]
- Chang, Z.K.; Meng, F.G.; Zhang, Z.Q.; Mao, G.P.; Huang, Z.Y.; Liao, W.M.; He, A.S. microRNA-193b-3p regulates matrix metalloproteinase 19 expression in interleukin-1beta-induced human chondrocytes. J. Cell. Biochem. 2018, 119, 4775–4782. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, R.; Zhang, Q.; Xu, Z.; Xu, F.; Li, D.; Li, Y. Chondrocyte sheet in vivo cartilage regeneration technique using miR-193b-3p to target MMP16. Aging 2019, 11, 7070–7082. [Google Scholar] [CrossRef]
- Xu, L.; Cao, H.; Xie, Y.; Zhang, Y.; Du, M.; Xu, X.; Ye, R.; Liu, X. Exosome-shuttled miR-92b-3p from ischemic preconditioned astrocytes protects neurons against oxygen and glucose deprivation. Brain Res. 2019, 1717, 66–73. [Google Scholar] [CrossRef]
- Hao, X.; Ma, C.; Chen, S.; Dang, J.; Cheng, X.; Zhu, D. Reverse the down regulation of miR-92b-3p by hypoxia can suppress the proliferation of pulmonary artery smooth muscle cells by targeting USP28. Biochem. Biophys. Res. Commun. 2018, 503, 3064–3077. [Google Scholar] [CrossRef]
- Li, T.; Liu, X.; Gong, X.; E, Q.; Zhang, X.; Zhang, X. microRNA 92b-3p regulates primordial follicle assembly by targeting TSC1 in neonatal mouse ovaries. Cell. Cycle 2019, 18, 824–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Z.; Liu, B.; Li, Y.; Liu, F.; Yuan, X.; Wang, Y. MicroRNA-652-3p promotes the proliferation and invasion of the trophoblast HTR-8/SVneo cell line by targeting homeobox A9 to modulate the expression of ephrin receptor B4. Clin. Exp. Pharm. Physiol. 2019, 46, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Qi, X.; Gui, Y.; Huo, H.; Yang, X.; Yang, L. Overexpression of circ_0021093 circular RNA forecasts an unfavorable prognosis and facilitates cell progression by targeting the miR-766-3p/MTA3 pathway in hepatocellular carcinoma. Gene 2019, 714, 143992. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, K.; Kawasaki, M.; Hirai, T.; Yoshida, Y.; Tsushima, H.; Fujishiro, M.; Ikeda, K.; Morimoto, S.; Takamori, K.; Sekigawa, I. MicroRNA-766-3p contributes to anti-inflammatory responses through the indirect inhibition of NF-κB signaling. Int. J. Mol. Sci. 2019, 20, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Biological Processes | Ribosomal Proteins (RPs) Involved |
|---|---|
| 1. SRP-dependent cotranslational protein targeting to membrane (GO:0006614) | RPL4, RPL5, RPL30, RPL3, RPL32, RPL31, RPL34, RPLP1, RPLP0, RPL10A, RPL8, RPL9, RPL6, RPL7, RPS4X, RPS15, RPS14, RPL7A, RPS17, RPS16, RPS19, RPL18A, RPS18, RPL36, RPLP2, RPL35, RPL37, RPS11, RPL39, RPS10, RPS13, RPS12, RPS9, RPL21, RPS7, RPS8, RPL23, RPS5, RPL22, RPS6, RPL13A, RPS3A, RPSA, RPL24, RPL27, RPL26, UBA52, RPL10, RPL12, RPL36A, RPS4Y1, RPS15A, RPS3, RPL14, RPS2, RPL15, RPS27A, RPL18, RPL17, RPL19, RPL41, RPL23A, RPS26, RPS25, RPS28, RPS27, RPS29, RPL27A, RPS20, FAU, RPS21, RPS24, RPS23 |
| 2. Viral transcription (GO:0019083) | |
| 3. Nuclear-transcribed mRNA catabolic process, nonsense-mediated decay (GO:0000184) | |
| 4. Translational initiation (GO:0006413) | |
| 5. rRNA processing (GO:0006364) | |
| 6. Translation (GO:0006412) | |
| 7. Cytoplasmic translation (GO:0002181) | RPL31, RPLP1, RPL22, RPLP0, RPL36A, RPL8, RPL9, RPL6, RPL7, RPL36, RPLP2, RPL26, RPL15 |
| 8. Ribosomal small subunit assembly (GO:0000028) | RPS15, RPS14, RPS17, RPS28, RPS27, RPS19, RPS5, RPSA, RPS10 |
| 9. Ribosomal small subunit biogenesis (GO:0042274) | RPS15, RPS17, RPS28, RPS16, RPS7, RPS19, RPS6, RPS24 |
| 10. Ribosomal large subunit assembly (GO:0000027) | RPL5, RPL3, RPL10, RPL12, RPL24, RPL23A, RPL6 |
| 11. Maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) (GO:0000462) | RPS14, RPS16, RPS19, RPS8, RPS24 |
| 12. Ribosome biogenesis (GO:0042254) | RPL7A, RPS28, RPS18, RPL34, RPLP0 |
| 13. Cell-cell adhesion (GO:0098609) | RPS26, RPL7A, RPL34, RPL14, RPL24, RPL23A, RPL15, RPS2, RPL6 |
| 14. Ribosomal large subunit biogenesis (GO:0042273) | RPL5, RPL14, RPL26, RPL7 |
| 15. Liver regeneration (GO:0097421) | RPS16, RPL32, RPS24, RPL19 |
| 16. Maturation of SSU-rRNA (GO:0030490) | RPS14, RPS28, RPS19 |
| 17. Endonucleolytic cleavage to generate mature 3′-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) (GO:0000461) | RPSA, RPS21 |
| 18. DNA damage response, detection of DNA damage (GO:0042769) | RPS3, RPS27A, UBA52 |
| 19. Negative regulation of RNA splicing (GO:0033119) | RPS26, RPS13 |
| 20. Endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) (GO:0000447) | RPSA, RPS21 |
| 21. Maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) (GO:0000463) | RPL35, RPL7 |
| 22. Erythrocyte homeostasis (GO:0034101) | RPS17, RPS24 |
| 23. Regulation of necrotic cell death (GO:0010939) | RPS27A, UBA52 |
| 24. Virion assembly (GO:0019068) | RPS27A, UBA52 |
| 25. Regulation of type I interferon production (GO:0032479) | RPS27A, UBA52 |
| 26. MyD88-independent toll-like receptor signaling pathway (GO:0002756) | RPS27A, UBA52 |
| 27. Maturation of LSU-rRNA (GO:0000470) | RPL7A, RPL10A |
| 28. Translational elongation (GO:0006414) | RPLP1, RPLP2 |
| 29. Response to ethanol (GO:0045471) | RPS4X, RPL15, RPL10A |
| 30. Error-free translesion synthesis (GO:0070987) | RPS27A, UBA52 |
| 31. Error-prone translesion synthesis (GO:0042276) | RPS27A, UBA52 |
| 32. Stress-activated MAPK cascade (GO:0051403) | RPS27A, UBA52 |
| 33. Notch signaling pathway (GO:0007219) | RPS19, RPS27A, UBA52 |
| 34. Nucleotide-excision repair, DNA duplex unwinding (GO:0000717) | RPS27A, UBA52 |
| 35. Nucleotide-excision repair, DNA damage recognition (GO:0000715) | RPS27A, UBA52 |
| 36. Positive regulation of epidermal growth factor receptor signaling pathway (GO:0045742) | RPS27A, UBA52 |
| 37. Nucleotide-excision repair, DNA gap filling (GO:0006297) | RPS27A, UBA52 |
| 38. Cellular response to interleukin-4 (GO:0071353) | RPL3, RPLP0 |
| Ribosomal Proteins | Functions | References |
|---|---|---|
| RPL33 | Regulates the processing of the 35S and 27S pre-rRNAs | [28] |
| RPS20 | Regulates mRNA binding and subunits association, mutation impairs 70S subunit formation and mRNA binding to the 30S subunit | [30] |
| RPL16 | Assembly of 60S subunits | [31,52] |
| RP59 | Assembly of the 40S subunit | [31] |
| RPL1 | Maintain the stability of 5S rRNA and assembly of 60S subunits | [32,33] |
| RPL9 | Maturation of the small subunit | [34] |
| RPL23 | Chaperone-assisted folding of proteins | [37] |
| RPL35 | Recognition of peptide and insertion to the translocation channel | [38] |
| RPS12 | Mutations at lysine-42 of S12, increase accuracy of translation | [41] |
| RPS4 | Mutation reduces the accuracy of translation | [41,43] |
| RPS5 | Mutation reduces the accuracy of translation | [41] |
| RPS9 | Maintain the accuracy of translation | [42] |
| RPS28 | Maintain the accuracy of translation | [43] |
| RPL39 | Maintain the accuracy of translation | [61] |
| RPL3 | Regulates the peptidyltransferase activity and mutation alter the fidelity of translation | [45] |
| RPL5 | Regulates the peptidyltransferase by helping the anchor of peptidyl-tRNA to the P-site | [47] |
| RPL41 | Optimizes peptidyltransferase activity by regulating the translocation | [46] |
| RPL24 | Regulates the P-site binding and kinetics of the protein synthesis | [61] |
| RPL10 | Regulates nuclear exporting by interacting and releasing cytoplasmic Nmd3p from 60S subunit | [48,49,50] |
| RPL12 | Assembly of ribosomal stalk | [51] |
| RPS14 | Maturation of 43S preribosomes | [53] |
| RPS0 | 20S rRNA-precursor to mature 18S rRNA | [54,55] |
| RPS21 | Maturation of the 3′ end of 18S rRNA | [55] |
| RPL25 | Pre-rRNA processing | [56] |
| RPS15 | Regulates the nuclear exit of the 40S subunit precursors | [57] |
| Major Ribosome Related Diseases | Gene Involved | Reference |
|---|---|---|
| Diamond–Blackfan anemia (DBA) | RPS19, RPS26, RPL5, RPL11 | [70,71,72] |
| 5q-syndrome | RPS14 | [73] |
| Schwachman-Diamond syndrome (SDS) | SBDS | [74] |
| X-linked dyskeratosis congenita (DC) | DKC1 | [75] |
| Cartilage hair hypoplasia (CHH) | RMRP | [76] |
| Treacher Collins syndrome (TCS) | TCOF1 | [77] |
| Bowen–Conradi syndrome (BCS) | EMG1 | [78,79,80] |
| North American Indian childhood cirrhosis (NAIC) | CIRH1A | [81,82] |
| Non-small cell lung cancer | RPS6, RPS15A | [83,84] |
| Ovarian cancer | RPS4X | [85] |
| Bladder cancer | RPS4X | [86] |
| Prostate cancer | RPL31 | [87] |
| Esophageal cancer | RPL34 | [88] |
| T-cell lymphoma | RPL22 | [89] |
| Colorectal cancer | RPS20 | [90] |
| miRNA | Predicted Target Ribosomal Proteins | Reported Disease Association |
|---|---|---|
| hsa-mir-16-5p | RPSA, RPL4, RPL9, RPL30, RPS3A, RPS15A, RPS24, RPL12, RPL3, RPL31, RPS6, RPL10, RPL27A, RPLP2, RPS27, RPS5, RPL14, RPL36, RPL5, RPL6, RPL21, RPLP0, RPLP1, RPS2, RPS3, RPS12, RPS17, RPS25, RPS14, RPL19, RPL15, RPL35, RPS19, RPS11 | breast cancer [147,148], hepatocellular carcinoma (HCC) [149], mesothelioma [150], glioma [151], neuroblastoma [152], chordoma, gastric cancer and osteoarthritis [153,154,155], osteoclastogenesis [156], and rheumatoid arthritis [157]. |
| hsa-mir-92a-3p | RPL9, RPL30, RPS3A, RPS10, RPS15A, RPS24, RPL18A, RPL3, RPL27A, RPS5, RPLP1, RPS25, RPL37, RPL7A, RPL24, FAU, RPS14, RPS28, RPL23, RPL13A, RPL39, RPL22, RPS23, RPL8, RPL15, RPL27, RPS8, RPS15 | melanoma [158], liposarcoma [159], glioma [160], leukemia [161], colorectal cancer [162,163], degradation of cartilage [164], Kawasaki disease [165], schizophrenia [166], systemic lupus erythematosus [167], and white matter impairment and post-stroke depression [168]. |
| hsa-mir-100-5p | RPS15A, RPL12, RPL31, RPL10, RPS27, RPL14, RPL5, RPL21, RPLP1, RPS2, RPL7, RPL36A, RPL7A, RPL26, RPL19, RPL15, RPS8, RPS15, RPL10A | prostate cancer [169], RCC [170], oral cancer [171], NSCLC [172], HCC [173], abeta-induced pathologies [174], and hidradenitis suppurativa [175]. |
| hsa-mir-615-3p | RPSA, RPL9, RPL3, UBA52, RPL31, RPL36, RPL5, RPL21, RPLP1, RPS2, RPS3, RPS12, RPS17, RPL7, RPL7A, RPL23, RPL15, RPS15 | gastric cancer [176], prostate cancer [177], NSCLC [178,179], esophageal cancer [180], and HCC [181]. |
| hsa-mir-484 | RPSA, RPS10, RPS24, RPL3, UBA52, RPL27A, RPLP2, RPL36, RPL5, RPL21, RPLP0, RPLP1, RPS3, FAU, RPS14, RPL13A, RPS18, RPS23, RPS15, RPS29, RPS9, RPL23A | cervical cancer [182,183], gastric cancer [184], glioma [185], NSCLC [186], adrenocortical cancer [187], colorectal cancer [188], renal carcinoma [189], and breast cancer [190]. |
| hsa-mir-186-5p | RPL4, RPL9, RPS4X, RPL18A, RPL3, RPL14, RPL36, RPL5, RPLP0, RPLP1, RPS2, RPS3, RPS7, RPS26, RPL15, RPL27, RPS29, RPL32, RPS21 | ischemia stroke [191], hippocampal neurons [192], acute coronary syndrome [193], reproductive health [194], osteosarcoma [195,196], colorectal cancer [197], NSCLC and neuroblastoma [198], lung adenocarcinoma [199], and prostate cancer [200]. |
| hsa-mir-320a | RPL9, RPL30, RPS4X, UBA52, RPL10, RPS27, RPL36, RPLP1, RPS2, RPS12, RPS17, RPS16, RPL7A, RPL13A, RPL8, RPL15, RPL27 | HCC [201,202], NSCLC [203], gliomas [204], gastric cancer [205,206], lung adenocarcinoma [207], tongue squamous cell carcinoma [208], multiple myeloma [209], breast cancer [210], colorectal cancer [211], bladder carcinoma [212], diabetic nephropathy [213], cartilage degradation [214], osteoporosis [215,216], cardiotoxicity [217], anomalous placentation [218], atherogenesis [219], arrhythmogenic cardiomyopathy [220], and polycystic ovary syndrome [221]. |
| hsa-mir-193b-3p | RPL9, RPS10, RPL12, RPS6, RPL27A, RPL6, RPLP0, RPS3, FAU, RPL26, RPS18, RPL22, RPL8, RPS21, RPL23A | preeclampsia [222], ovarian cancer [223], breast cancer [224], and urothelial cancer [225]. |
| hsa-let-7a-5p | RPSA, RPL4, RPL9, RPL30, RPS3A, RPS4X, RPS10, RPS13, RPS15A, RPS24, RPL31, RPS27, RPLP1, RPS14, RPL8, RPS29 | osteogenesis [226], lung cancer [227,228], HCC [229], colorectal cancer [230], leukemia cells [231], diabetic nephropathy [232], hepatic fibrosis [233]. |
| hsa-mir-331-3p | RPS4Y1, RPS27, RPLP0, RPLP1, RPS2, RPS3, RPS12, RPL36A, RPL7A, RPS14, RPL13A, RPS29, RPS9, RPL34 | pancreatic cancer [234], HCC [235,236,237], esophageal adenocarcinoma [238], prostate cancer [239,240], colorectal cancer [241], NSCLC [242], glioblastoma and cervical cancer [243,244], ovarian cancer [245], urothelial cancer [246], and gastric cancer [247], intracranial aneurysm [248]. |
| hsa-mir-92b-3p | RPL9, RPL30, RPS3A, RPS4X, RPL3, RPLP0, RPL37, RPL24, RPS14, RPS28, RPL23, RPL8 | acute spinal cord injury [249], cardiac hypertrophy [250,251], pancreatic cancer [252], colorectal cancer [253], esophageal squamous cell carcinoma [254], gastric cancer [255], and synovial sarcoma [256]. |
| hsa-mir-652-3p | RPL4, RPL18, RPL18A, RPS6, RPL21, RPLP1, RPS12, RPS16, RPL26, RPL19, RPS23, RPL27, RPS29, RPL32, RPS19 | lymphoblastic leukemia [257], bladder cancer [258], NSCLC [259], prostate cancer [260], and atherosclerosis [261]. |
| hsa-mir-766-3p | RPS15A, RPL12, UBA52, RPL27A, RPLP2, RPL23, RPS26, RPL27, RPS9, RPS19, RPS21 | HCC [262], and RCC [263]. |
| hsa-mir-744-5p | RPL4, RPL18, RPL18A, RPL3, RPS6, RPL10, RPL36, RPLP1, RPL37, RPL7A, RPS14 | ovarian cancer [264], and NSCLC [265]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reza, A.M.M.T.; Yuan, Y.-G. microRNAs Mediated Regulation of the Ribosomal Proteins and its Consequences on the Global Translation of Proteins. Cells 2021, 10, 110. https://doi.org/10.3390/cells10010110
Reza AMMT, Yuan Y-G. microRNAs Mediated Regulation of the Ribosomal Proteins and its Consequences on the Global Translation of Proteins. Cells. 2021; 10(1):110. https://doi.org/10.3390/cells10010110
Chicago/Turabian StyleReza, Abu Musa Md Talimur, and Yu-Guo Yuan. 2021. "microRNAs Mediated Regulation of the Ribosomal Proteins and its Consequences on the Global Translation of Proteins" Cells 10, no. 1: 110. https://doi.org/10.3390/cells10010110
APA StyleReza, A. M. M. T., & Yuan, Y. -G. (2021). microRNAs Mediated Regulation of the Ribosomal Proteins and its Consequences on the Global Translation of Proteins. Cells, 10(1), 110. https://doi.org/10.3390/cells10010110