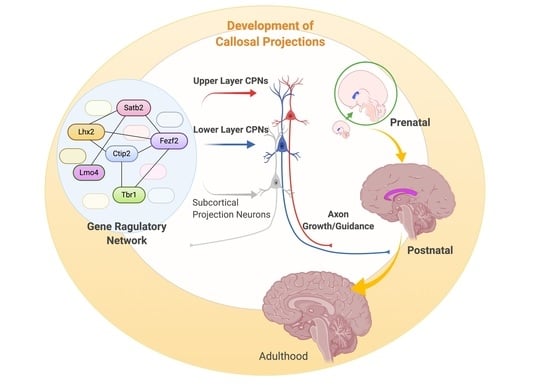
New Molecular Players in the Development of Callosal Projections
Abstract
:1. Introduction—The Corpus Callosum
2. Prenatal Development of Callosal Projections
2.1. Specification of Callosal Projection Neurons
2.1.1. SATB2-mediated Specification
2.1.2. Other Players in CPN Specification
2.2. Guidance of Callosal Axons
2.2.1. Players in Semaphorin/Neuropilin/Plexin Pathway
2.2.2. Players in Slit/Robo Pathway
2.2.3. Players in Netrin/DCC/Unc5 Pathway
3. Postnatal Development of Callosal Projections
3.1. Postnatal Specification of Callosal Projection Neurons
3.1.1. Transcription Factors
3.1.2. Other Players in CPN Specification
3.2. Callosal Axon Guidance During the Postnatal Period
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paul, L.K.; Brown, W.S.; Adolphs, R.; Tyszka, J.M.; Richards, L.J.; Mukherjee, P.; Sherr, E.H. Agenesis of the corpus callosum: Genetic, developmental and functional aspects of connectivity. Nat. Rev. Neurosci. 2007, 8, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Paul, L.K. Developmental malformation of the corpus callosum: A review of typical callosal development and examples of developmental disorders with callosal involvement. J. Neurodev. Disord 2011, 3, 3–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.J.; Kim, J.J.; Lee, S.K.; Seok, J.H.; Chun, J.; Kim, D.I.; Lee, J.D. Corpus callosal connection mapping using cortical gray matter parcellation and DT-MRI. Hum. Brain Mapp. 2008, 29, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Putnam, M.C.; Steven, M.S.; Doron, K.W.; Riggall, A.C.; Gazzaniga, M.S. Cortical projection topography of the human splenium: Hemispheric asymmetry and individual differences. J. Cogn. Neurosci. 2010, 22, 1662–1669. [Google Scholar] [CrossRef]
- Wahl, M.; Lauterbach-Soon, B.; Hattingen, E.; Jung, P.; Singer, O.; Volz, S.; Klein, J.C.; Steinmetz, H.; Ziemann, U. Human motor corpus callosum: Topography, somatotopy, and link between microstructure and function. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 12132–12138. [Google Scholar] [CrossRef] [Green Version]
- Funnell, M.G.; Corballis, P.M.; Gazzaniga, M.S. Cortical and subcortical interhemispheric interactions following partial and complete callosotomy. Arch. Neurol. 2000, 57, 185–189. [Google Scholar] [CrossRef]
- Blaauw, J.; Meiners, L.C. The splenium of the corpus callosum: Embryology, anatomy, function and imaging with pathophysiological hypothesis. Neuroradiology 2020, 62, 563–585. [Google Scholar] [CrossRef] [Green Version]
- Niquille, M.; Garel, S.; Mann, F.; Hornung, J.P.; Otsmane, B.; Chevalley, S.; Parras, C.; Guillemot, F.; Gaspar, P.; Yanagawa, Y.; et al. Transient neuronal populations are required to guide callosal axons: A role for semaphorin 3C. PLoS Biol. 2009, 7, e1000230. [Google Scholar] [CrossRef] [Green Version]
- Nishikimi, M.; Oishi, K.; Tabata, H.; Torii, K.; Nakajima, K. Segregation and pathfinding of callosal axons through EphA3 signaling. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 16251–16260. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wen, Y.; She, L.; Sui, Y.N.; Liu, L.; Richards, L.J.; Poo, M.M. Axon position within the corpus callosum determines contralateral cortical projection. Proc. Natl. Acad. Sci. USA 2013, 110, E2714–E2723. [Google Scholar] [CrossRef] [Green Version]
- Fame, R.M.; MacDonald, J.L.; Macklis, J.D. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011, 34, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meissirel, C.; Dehay, C.; Berland, M.; Kennedy, H. Segregation of callosal and association pathways during development in the visual cortex of the primate. J. Neurosci. Off. J. Soc. Neurosci. 1991, 11, 3297–3316. [Google Scholar] [CrossRef] [Green Version]
- Wise, S.P.; Jones, E.G. The organization and postnatal development of the commissural projection of the rat somatic sensory cortex. J. Comp. Neurol. 1976, 168, 313–343. [Google Scholar] [CrossRef] [PubMed]
- Schulte, T.; Müller-Oehring, E.M. Contribution of callosal connections to the interhemispheric integration of visuomotor and cognitive processes. Neuropsychol. Rev. 2010, 20, 174–190. [Google Scholar] [CrossRef] [Green Version]
- Forster, B.; Corballis, M.C. Interhemispheric transmission times in the presence and absence of the forebrain commissures: Effects of luminance and equiluminance. Neuropsychologia 1998, 36, 925–934. [Google Scholar] [CrossRef]
- Koshiyama, D.; Fukunaga, M.; Okada, N.; Morita, K.; Nemoto, K.; Yamashita, F.; Yamamori, H.; Yasuda, Y.; Fujimoto, M.; Kelly, S.; et al. Role of frontal white matter and corpus callosum on social function in schizophrenia. Schizophr. Res. 2018, 202, 180–187. [Google Scholar] [CrossRef]
- Shafer, A.T.; Benoit, J.R.; Brown, M.R.G.; Greenshaw, A.J.; Van Vliet, K.J.; Vohra, S.; Dolcos, F.; Singhal, A. Differences in attentional control and white matter microstructure in adolescents with attentional, affective, and behavioral disorders. Brain Imaging Behav. 2020, 14, 599–614. [Google Scholar] [CrossRef]
- Jiang, W.; Shi, F.; Liu, H.; Li, G.; Ding, Z.; Shen, H.; Shen, C.; Lee, S.W.; Hu, D.; Wang, W.; et al. Reduced White Matter Integrity in Antisocial Personality Disorder: A Diffusion Tensor Imaging Study. Sci. Rep. 2017, 7, 43002. [Google Scholar] [CrossRef] [Green Version]
- Mathews, M.S.; Linskey, M.E.; Binder, D.K. William P. van Wagenen and the first corpus callosotomies for epilepsy. J. Neurosurg. 2008, 108, 608–613. [Google Scholar] [CrossRef] [Green Version]
- Barkovich, A.J.; Norman, D. Anomalies of the corpus callosum: Correlation with further anomalies of the brain. Ajr. Am. J. Roentgenol. 1988, 151, 171–179. [Google Scholar] [CrossRef] [Green Version]
- Richards, L.J.; Plachez, C.; Ren, T. Mechanisms regulating the development of the corpus callosum and its agenesis in mouse and human. Clin. Genet. 2004, 66, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Kale, A.; Joshi, P.; Kelkar, A.B. Restricted diffusion in the corpus callosum: A neuroradiological marker in hypoxic-ischemic encephalopathy. Indian J. Radiol. Imaging 2016, 26, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Jarmasz, J.S.; Basalah, D.A.; Chudley, A.E.; Del Bigio, M.R. Human brain abnormalities associated with prenatal alcohol exposure and fetal alcohol spectrum disorder. J. Neuropathol. Exp. Neurol. 2017, 76, 813–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, D.; Tisdall, M.M.; Gill, D. Corpus callosotomy outcomes in pediatric patients: A systematic review. Epilepsia 2016, 57, 1053–1068. [Google Scholar] [CrossRef] [PubMed]
- Koester, S.E.; O’Leary, D.D. Connectional distinction between callosal and subcortically projecting cortical neurons is determined prior to axon extension. Dev. Biol. 1993, 160, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, H.S.; Wahlsten, D. Prenatal formation of the normal mouse corpus callosum: A quantitative study with carbocyanine dyes. J. Comp. Neurol. 1992, 323, 81–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rash, B.G.; Richards, L.J. A role for cingulate pioneering axons in the development of the corpus callosum. J. Comp. Neurol. 2001, 434, 147–157. [Google Scholar] [CrossRef]
- Rakic, P.; Yakovlev, P.I. Development of the corpus callosum and cavum septi in man. J. Comp. Neurol. 1968, 132, 45–72. [Google Scholar] [CrossRef]
- Cignini, P.; Padula, F.; Giorlandino, M.; Brutti, P.; Alfò, M.; Giannarelli, D.; Mastrandrea, M.L.; D’Emidio, L.; Vacca, L.; Aloisi, A.; et al. Reference charts for fetal corpus callosum length: A prospective cross-sectional study of 2950 fetuses. J. Ultrasound Med. Off. J. Am. Inst. Ultrasound Med. 2014, 33, 1065–1078. [Google Scholar] [CrossRef]
- Horgos, B.; Mecea, M.; Boer, A.; Szabo, B.; Buruiana, A.; Stamatian, F.; Mihu, C.-M.; Florian, I.Ş.; Susman, S.; Pascalau, R. White Matter Dissection of the Fetal Brain. Front. Neuroanat. 2020, 14. [Google Scholar] [CrossRef]
- Achiron, R.; Achiron, A. Development of the human fetal corpus callosum: A high-resolution, cross-sectional sonographic study. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2001, 18, 343–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harreld, J.H.; Bhore, R.; Chason, D.P.; Twickler, D.M. Corpus callosum length by gestational age as evaluated by fetal MR imaging. AJNR Am. J. Neuroradiol. 2011, 32, 490–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De León Reyes, N.S.; Bragg-Gonzalo, L.; Nieto, M. Development and plasticity of the corpus callosum. Development 2020, 147, dev189738. [Google Scholar] [CrossRef] [PubMed]
- Raybaud, C. The corpus callosum, the other great forebrain commissures, and the septum pellucidum: Anatomy, development, and malformation. Neuroradiology 2010, 52, 447–477. [Google Scholar] [CrossRef]
- Molyneaux, B.J.; Arlotta, P.; Menezes, J.R.; Macklis, J.D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007, 8, 427–437. [Google Scholar] [CrossRef]
- Klingler, E.; De la Rossa, A.; Fièvre, S.; Devaraju, K.; Abe, P.; Jabaudon, D. A Translaminar Genetic Logic for the Circuit Identity of Intracortically Projecting Neurons. Curr. Biol. 2019, 29, 332–339.e335. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, H.S.; Wahlsten, D. Timing and origin of the first cortical axons to project through the corpus callosum and the subsequent emergence of callosal projection cells in mouse. J. Comp. Neurol. 1998, 400, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Leone, D.P.; Srinivasan, K.; Chen, B.; Alcamo, E.; McConnell, S.K. The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol. 2008, 18, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Leyva-Díaz, E.; López-Bendito, G. In and out from the cortex: Development of major forebrain connections. Neuroscience 2013, 254, 26–44. [Google Scholar] [CrossRef]
- Muralidharan, B.; Khatri, Z.; Maheshwari, U.; Gupta, R.; Roy, B.; Pradhan, S.J.; Karmodiya, K.; Padmanabhan, H.; Shetty, A.S.; Balaji, C.; et al. LHX2 Interacts with the NuRD Complex and Regulates Cortical Neuron Subtype Determinants Fezf2 and Sox11. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Shim, S.; Kwan, K.Y.; Li, M.; Lefebvre, V.; Sestan, N. Cis-regulatory control of corticospinal system development and evolution. Nature 2012, 486, 74–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwan, K.Y.; Lam, M.M.; Krsnik, Z.; Kawasawa, Y.I.; Lefebvre, V.; Sestan, N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. USA 2008, 105, 16021–16026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcamo, E.A.; Chirivella, L.; Dautzenberg, M.; Dobreva, G.; Fariñas, I.; Grosschedl, R.; McConnell, S.K. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 2008, 57, 364–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britanova, O.; de Juan Romero, C.; Cheung, A.; Kwan, K.Y.; Schwark, M.; Gyorgy, A.; Vogel, T.; Akopov, S.; Mitkovski, M.; Agoston, D.; et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 2008, 57, 378–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paolino, A.; Fenlon, L.R.; Kozulin, P.; Haines, E.; Lim, J.W.C.; Richards, L.J.; Suárez, R. Differential timing of a conserved transcriptional network underlies divergent cortical projection routes across mammalian brain evolution. Proc. Natl. Acad. Sci. USA 2020, 117, 10554–10564. [Google Scholar] [CrossRef]
- Baranek, C.; Dittrich, M.; Parthasarathy, S.; Bonnon, C.G.; Britanova, O.; Lanshakov, D.; Boukhtouche, F.; Sommer, J.E.; Colmenares, C.; Tarabykin, V.; et al. Protooncogene Ski cooperates with the chromatin-remodeling factor Satb2 in specifying callosal neurons. Proc. Natl. Acad. Sci. USA 2012, 109, 3546–3551. [Google Scholar] [CrossRef] [Green Version]
- Harb, K.; Magrinelli, E.; Nicolas, C.S.; Lukianets, N.; Frangeul, L.; Pietri, M.; Sun, T.; Sandoz, G.; Grammont, F.; Jabaudon, D.; et al. Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. eLife 2016, 5, e09531. [Google Scholar] [CrossRef]
- McKenna, W.L.; Ortiz-Londono, C.F.; Mathew, T.K.; Hoang, K.; Katzman, S.; Chen, B. Mutual regulation between Satb2 and Fezf2 promotes subcerebral projection neuron identity in the developing cerebral cortex. Proc. Natl. Acad. Sci. USA 2015, 112, 11702–11707. [Google Scholar] [CrossRef] [Green Version]
- Paşca, S.P.; Portmann, T.; Voineagu, I.; Yazawa, M.; Shcheglovitov, A.; Paşca, A.M.; Cord, B.; Palmer, T.D.; Chikahisa, S.; Nishino, S.; et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat. Med. 2011, 17, 1657–1662. [Google Scholar] [CrossRef]
- Panagiotakos, G.; Haveles, C.; Arjun, A.; Petrova, R.; Rana, A.; Portmann, T.; Paşca, S.P.; Palmer, T.D.; Dolmetsch, R.E. Aberrant calcium channel splicing drives defects in cortical differentiation in Timothy syndrome. eLife 2019, 8, e51037. [Google Scholar] [CrossRef]
- Büttner, N.; Johnsen, S.A.; Kügler, S.; Vogel, T. Af9/Mllt3 interferes with Tbr1 expression through epigenetic modification of histone H3K79 during development of the cerebral cortex. Proc. Natl. Acad. Sci. USA 2010, 107, 7042–7047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franz, H.; Villarreal, A.; Heidrich, S.; Videm, P.; Kilpert, F.; Mestres, I.; Calegari, F.; Backofen, R.; Manke, T.; Vogel, T. DOT1L promotes progenitor proliferation and primes neuronal layer identity in the developing cerebral cortex. Nucleic Acids Res. 2019, 47, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Saavedra, M.; Yan, K.; De Repentigny, Y.; Hashem, L.E.; Chaudary, N.; Sarwar, S.; Yang, D.; Ioshikhes, I.; Kothary, R.; Hirayama, T.; et al. Snf2h Drives Chromatin Remodeling to Prime Upper Layer Cortical Neuron Development. Front. Mol. Neurosci. 2019, 12, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, L.; Kim, N.H.; Huh, S.O.; Rhee, H.J. Depletion of Inositol Polyphosphate 4-Phosphatase II Suppresses Callosal Axon Formation in the Developing Mice. Mol. Cells 2016, 39, 501–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatanaka, Y.; Namikawa, T.; Yamauchi, K.; Kawaguchi, Y. Cortical Divergent Projections in Mice Originate from Two Sequentially Generated, Distinct Populations of Excitatory Cortical Neurons with Different Initial Axonal Outgrowth Characteristics. Cereb. Cortex 2016, 26, 2257–2270. [Google Scholar] [CrossRef] [Green Version]
- Srivatsa, S.; Parthasarathy, S.; Britanova, O.; Bormuth, I.; Donahoo, A.L.; Ackerman, S.L.; Richards, L.J.; Tarabykin, V. Unc5C and DCC act downstream of Ctip2 and Satb2 and contribute to corpus callosum formation. Nat. Commun. 2014, 5, 3708. [Google Scholar] [CrossRef] [Green Version]
- Marsh, A.P.; Heron, D.; Edwards, T.J.; Quartier, A.; Galea, C.; Nava, C.; Rastetter, A.; Moutard, M.L.; Anderson, V.; Bitoun, P.; et al. Mutations in DCC cause isolated agenesis of the corpus callosum with incomplete penetrance. Nat Genet 2017, 49, 511–514. [Google Scholar] [CrossRef]
- Srinivasan, K.; Leone, D.P.; Bateson, R.K.; Dobreva, G.; Kohwi, Y.; Kohwi-Shigematsu, T.; Grosschedl, R.; McConnell, S.K. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc. Natl. Acad. Sci. USA 2012, 109, 19071–19078. [Google Scholar] [CrossRef] [Green Version]
- Greig, L.C.; Woodworth, M.B.; Galazo, M.J.; Padmanabhan, H.; Macklis, J.D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 2013, 14, 755–769. [Google Scholar] [CrossRef] [Green Version]
- Fame, R.M.; Dehay, C.; Kennedy, H.; Macklis, J.D. Subtype-Specific Genes that Characterize Subpopulations of Callosal Projection Neurons in Mouse Identify Molecularly Homologous Populations in Macaque Cortex. Cereb Cortex 2017, 27, 1817–1830. [Google Scholar] [CrossRef] [Green Version]
- Heavner, W.E.; Ji, S.; Notwell, J.H.; Dyer, E.S.; Tseng, A.M.; Birgmeier, J.; Yoo, B.; Bejerano, G.; McConnell, S.K. Transcription factor expression defines subclasses of developing projection neurons highly similar to single-cell RNA-seq subtypes. Proc. Natl. Acad. Sci. USA 2020, 117, 25074–25084. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.L.; Fame, R.M.; Gillis-Buck, E.M.; Macklis, J.D. Caveolin1 Identifies a Specific Subpopulation of Cerebral Cortex Callosal Projection Neurons (CPN) Including Dual Projecting Cortical Callosal/Frontal Projection Neurons (CPN/FPN). eNeuro 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Molyneaux, B.J.; Arlotta, P.; Fame, R.M.; MacDonald, J.L.; MacQuarrie, K.L.; Macklis, J.D. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J. Neurosci. Off. J. Soc. Neurosci. 2009, 29, 12343–12354. [Google Scholar] [CrossRef] [PubMed]
- Molyneaux, B.J.; Goff, L.A.; Brettler, A.C.; Chen, H.H.; Hrvatin, S.; Rinn, J.L.; Arlotta, P. DeCoN: Genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 2015, 85, 275–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paolino, A.; Fenlon, L.R.; Suárez, R.; Richards, L.J. Transcriptional control of long-range cortical projections. Curr. Opin. Neurobiol. 2018, 53, 57–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cargnin, F.; Kwon, J.S.; Katzman, S.; Chen, B.; Lee, J.W.; Lee, S.K. FOXG1 Orchestrates Neocortical Organization and Cortico-Cortical Connections. Neuron 2018, 100, 1083–1096.e1085. [Google Scholar] [CrossRef] [Green Version]
- Kumamoto, T.; Toma, K.; Gunadi; McKenna, W.L.; Kasukawa, T.; Katzman, S.; Chen, B.; Hanashima, C. Foxg1 coordinates the switch from nonradially to radially migrating glutamatergic subtypes in the neocortex through spatiotemporal repression. Cell Rep. 2013, 3, 931–945. [Google Scholar] [CrossRef] [Green Version]
- Hou, P.S.; Miyoshi, G.; Hanashima, C. Sensory cortex wiring requires preselection of short- and long-range projection neurons through an Egr-Foxg1-COUP-TFI network. Nat. Commun. 2019, 10, 3581. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, S.A.; Bernard, A.; Menon, V.; Royall, J.J.; Glattfelder, K.J.; Desta, T.; Hirokawa, K.; Mortrud, M.; Miller, J.A.; Zeng, H.; et al. Correlated gene expression and target specificity demonstrate excitatory projection neuron diversity. Cereb Cortex 2015, 25, 433–449. [Google Scholar] [CrossRef] [Green Version]
- Tasic, B.; Menon, V.; Nguyen, T.N.; Kim, T.K.; Jarsky, T.; Yao, Z.; Levi, B.; Gray, L.T.; Sorensen, S.A.; Dolbeare, T.; et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016, 19, 335–346. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, C.K.; Beltramo, R. Correlating Anatomy and Function with Gene Expression in Individual Neurons by Combining in Vivo Labeling, Patch Clamp, and Single Cell RNA-seq. Front. Cell. Neurosci. 2017, 11, 376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, R.L.; Tam, D.M.; Brumberg, J.C. Physiology and morphology of callosal projection neurons in mouse. Neuroscience 2008, 153, 654–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azim, E.; Shnider, S.J.; Cederquist, G.Y.; Sohur, U.S.; Macklis, J.D. Lmo4 and Clim1 progressively delineate cortical projection neuron subtypes during development. Cereb Cortex 2009, 19 (Suppl. S1), i62–i69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cederquist, G.Y.; Azim, E.; Shnider, S.J.; Padmanabhan, H.; Macklis, J.D. Lmo4 establishes rostral motor cortex projection neuron subtype diversity. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 6321–6332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashani, A.H.; Qiu, Z.; Jurata, L.; Lee, S.K.; Pfaff, S.; Goebbels, S.; Nave, K.A.; Ghosh, A. Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 8398–8408. [Google Scholar] [CrossRef] [Green Version]
- Fame, R.M.; MacDonald, J.L.; Dunwoodie, S.L.; Takahashi, E.; Macklis, J.D. Cited2 Regulates Neocortical Layer II/III Generation and Somatosensory Callosal Projection Neuron Development and Connectivity. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 6403–6419. [Google Scholar] [CrossRef] [Green Version]
- Greig, L.C.; Woodworth, M.B.; Greppi, C.; Macklis, J.D. Ctip1 Controls Acquisition of Sensory Area Identity and Establishment of Sensory Input Fields in the Developing Neocortex. Neuron 2016, 90, 261–277. [Google Scholar] [CrossRef] [Green Version]
- Wiegreffe, C.; Simon, R.; Peschkes, K.; Kling, C.; Strehle, M.; Cheng, J.; Srivatsa, S.; Liu, P.; Jenkins, N.A.; Copeland, N.G.; et al. Bcl11a (Ctip1) Controls Migration of Cortical Projection Neurons through Regulation of Sema3c. Neuron 2015, 87, 311–325. [Google Scholar] [CrossRef] [Green Version]
- Woodworth, M.B.; Greig, L.C.; Liu, K.X.; Ippolito, G.C.; Tucker, H.O.; Macklis, J.D. Ctip1 Regulates the Balance between Specification of Distinct Projection Neuron Subtypes in Deep Cortical Layers. Cell Rep. 2016, 15, 999–1012. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Wang, S.S.; Hattox, A.M.; Rayburn, H.; Nelson, S.B.; McConnell, S.K. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc. Natl. Acad. Sci. USA 2008, 105, 11382–11387. [Google Scholar] [CrossRef] [Green Version]
- McKenna, W.L.; Betancourt, J.; Larkin, K.A.; Abrams, B.; Guo, C.; Rubenstein, J.L.; Chen, B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Molyneaux, B.J.; Arlotta, P.; Hirata, T.; Hibi, M.; Macklis, J.D. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron 2005, 47, 817–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Saavedra, M.; De Repentigny, Y.; Lagali, P.S.; Raghu Ram, E.V.; Yan, K.; Hashem, E.; Ivanochko, D.; Huh, M.S.; Yang, D.; Mears, A.J.; et al. Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nat. Commun. 2014, 5, 4181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazzaro, M.A.; Picketts, D.J. Cloning and characterization of the murine Imitation Switch (ISWI) genes: Differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem. 2001, 77, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Roidl, D.; Hellbach, N.; Bovio, P.P.; Villarreal, A.; Heidrich, S.; Nestel, S.; Grüning, B.A.; Boenisch, U.; Vogel, T. DOT1L Activity Promotes Proliferation and Protects Cortical Neural Stem Cells from Activation of ATF4-DDIT3-Mediated ER Stress In Vitro. Stem Cells (Dayt. Ohio) 2016, 34, 233–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, D.J.; Wilkinson, G.; Li, S.; Dixit, R.; Adnani, L.; Balakrishnan, A.; Han, S.; Kovach, C.; Gruenig, N.; Kurrasch, D.M.; et al. Neurog2 and Ascl1 together regulate a postmitotic derepression circuit to govern laminar fate specification in the murine neocortex. Proc. Natl. Acad. Sci. USA 2017, 114, e4934–e4943. [Google Scholar] [CrossRef] [Green Version]
- Fode, C.; Ma, Q.; Casarosa, S.; Ang, S.L.; Anderson, D.J.; Guillemot, F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000, 14, 67–80. [Google Scholar]
- Nieto, M.; Schuurmans, C.; Britz, O.; Guillemot, F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 2001, 29, 401–413. [Google Scholar] [CrossRef] [Green Version]
- Tao, W.; Lai, E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron 1992, 8, 957–966. [Google Scholar] [CrossRef]
- Izzi, L.; Charron, F. Midline axon guidance and human genetic disorders. Clin. Genet. 2011, 80, 226–234. [Google Scholar] [CrossRef]
- Suarez, R.; Gobius, I.; Richards, L.J. Evolution and development of interhemispheric connections in the vertebrate forebrain. Front. Hum. Neurosci. 2014, 8, 497. [Google Scholar] [CrossRef] [PubMed]
- De León Reyes, N.S.; Mederos, S.; Varela, I.; Weiss, L.A.; Perea, G.; Galazo, M.J.; Nieto, M. Transient callosal projections of L4 neurons are eliminated for the acquisition of local connectivity. Nat. Commun. 2019, 10, 4549. [Google Scholar] [CrossRef] [PubMed]
- Koester, S.E.; O’Leary, D.D. Axons of early generated neurons in cingulate cortex pioneer the corpus callosum. J. Neurosci. Off. J. Soc. Neurosci. 1994, 14, 6608–6620. [Google Scholar] [CrossRef] [Green Version]
- Choe, Y.; Siegenthaler, J.A.; Pleasure, S.J. A cascade of morphogenic signaling initiated by the meninges controls corpus callosum formation. Neuron 2012, 73, 698–712. [Google Scholar] [CrossRef] [Green Version]
- Hand, R.; Polleux, F. Neurogenin2 regulates the initial axon guidance of cortical pyramidal neurons projecting medially to the corpus callosum. Neural Dev. 2011, 6, 30. [Google Scholar] [CrossRef] [Green Version]
- Norris, C.R.; Kalil, K. Guidance of callosal axons by radial glia in the developing cerebral cortex. J. Neurosci. Off. J. Soc. Neurosci. 1991, 11, 3481–3492. [Google Scholar] [CrossRef] [Green Version]
- Shu, T.; Li, Y.; Keller, A.; Richards, L.J. The glial sling is a migratory population of developing neurons. Development 2003, 130, 2929–2937. [Google Scholar] [CrossRef] [Green Version]
- Shu, T.; Richards, L.J. Cortical axon guidance by the glial wedge during the development of the corpus callosum. J. Neurosci. Off. J. Soc. Neurosci. 2001, 21, 2749–2758. [Google Scholar] [CrossRef]
- Silver, J.; Edwards, M.A.; Levitt, P. Immunocytochemical demonstration of early appearing astroglial structures that form boundaries and pathways along axon tracts in the fetal brain. J. Comp. Neurol. 1993, 328, 415–436. [Google Scholar] [CrossRef]
- Silver, J.; Lorenz, S.E.; Wahlsten, D.; Coughlin, J. Axonal guidance during development of the great cerebral commissures: Descriptive and experimental studies, in vivo, on the role of preformed glial pathways. J. Comp. Neurol. 1982, 210, 10–29. [Google Scholar] [CrossRef]
- Hankin, M.H.; Schneider, B.F.; Silver, J. Death of the subcallosal glial sling is correlated with formation of the cavum septi pellucidi. J. Comp. Neurol. 1988, 272, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Unni, D.K.; Piper, M.; Moldrich, R.X.; Gobius, I.; Liu, S.; Fothergill, T.; Donahoo, A.L.; Baisden, J.M.; Cooper, H.M.; Richards, L.J. Multiple Slits regulate the development of midline glial populations and the corpus callosum. Dev. Biol. 2012, 365, 36–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, T.; Puche, A.C.; Richards, L.J. Development of midline glial populations at the corticoseptal boundary. J. Neurobiol. 2003, 57, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Niquille, M.; Minocha, S.; Hornung, J.P.; Rufer, N.; Valloton, D.; Kessaris, N.; Alfonsi, F.; Vitalis, T.; Yanagawa, Y.; Devenoges, C.; et al. Two specific populations of GABAergic neurons originating from the medial and the caudal ganglionic eminences aid in proper navigation of callosal axons. Dev. Neurobiol. 2013, 73, 647–672. [Google Scholar] [CrossRef] [Green Version]
- Jovanov-Milosevic, N.; Culjat, M.; Kostovic, I. Growth of the human corpus callosum: Modular and laminar morphogenetic zones. Front. Neuroanat. 2009, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Lindwall, C.; Fothergill, T.; Richards, L.J. Commissure formation in the mammalian forebrain. Curr. Opin. Neurobiol. 2007, 17, 3–14. [Google Scholar] [CrossRef]
- Wang, B.; Li, H.; Mutlu, S.A.; Bowser, D.A.; Moore, M.J.; Wang, M.C.; Zheng, H. The Amyloid Precursor Protein Is a Conserved Receptor for Slit to Mediate Axon Guidance. eNeuro 2017, 4. [Google Scholar] [CrossRef] [Green Version]
- DeGeer, J.; Kaplan, A.; Mattar, P.; Morabito, M.; Stochaj, U.; Kennedy, T.E.; Debant, A.; Cayouette, M.; Fournier, A.E.; Lamarche-Vane, N. Hsc70 chaperone activity underlies Trio GEF function in axon growth and guidance induced by netrin-1. J. Cell Biol. 2015, 210, 817–832. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.S.; Yang, Y.R.; Lee, C.; Park, B.; Park, K.I.; Seo, J.K.; Seo, Y.K.; Cho, H.; Lucio, C.; Suh, P.G. Netrin-1/DCC-mediated PLCγ1 activation is required for axon guidance and brain structure development. EMBO Rep. 2018, 19, e46250. [Google Scholar] [CrossRef]
- Brudvig, J.J.; Cain, J.T.; Schmidt-Grimminger, G.G.; Stumpo, D.J.; Roux, K.J.; Blackshear, P.J.; Weimer, J.M. MARCKS Is Necessary for Netrin-DCC Signaling and Corpus Callosum Formation. Mol. Neurobiol. 2018, 55, 8388–8402. [Google Scholar] [CrossRef]
- Weimer, J.M.; Yokota, Y.; Stanco, A.; Stumpo, D.J.; Blackshear, P.J.; Anton, E.S. MARCKS modulates radial progenitor placement, proliferation and organization in the developing cerebral cortex. Development 2009, 136, 2965–2975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez Juliá, A.; Frasch, A.C.; Fuchsova, B. Neuronal filopodium formation induced by the membrane glycoprotein M6a (Gpm6a) is facilitated by coronin-1a, Rac1, and p21-activated kinase 1 (Pak1). J. Neurochem. 2016, 137, 46–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mita, S.; de Monasterio-Schrader, P.; Fünfschilling, U.; Kawasaki, T.; Mizuno, H.; Iwasato, T.; Nave, K.A.; Werner, H.B.; Hirata, T. Transcallosal Projections Require Glycoprotein M6-Dependent Neurite Growth and Guidance. Cereb Cortex 2015, 25, 4111–4125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.M.; Tsuzuki, T.; Sakakibara, K.; Imaizumi, F.; Ikegaya, A.; Inagaki, M.; Takahashi, I.; Ito, T.; Takamatsu, H.; Kumanogoh, A.; et al. PlexinA1 is crucial for the midline crossing of callosal axons during corpus callosum development in BALB/cAJ mice. PLoS ONE 2019, 14, e0221440. [Google Scholar] [CrossRef]
- Wu, K.Y.; He, M.; Hou, Q.Q.; Sheng, A.L.; Yuan, L.; Liu, F.; Liu, W.W.; Li, G.; Jiang, X.Y.; Luo, Z.G. Semaphorin 3A activates the guanosine triphosphatase Rab5 to promote growth cone collapse and organize callosal axon projections. Sci. Signal. 2014, 7, ra81. [Google Scholar] [CrossRef] [Green Version]
- Velona, T.; Altounian, M.; Roque, M.; Hocine, M.; Bellon, A.; Briz, C.G.; Salin, P.; Nieto, M.; Chauvet, S.; Mann, F. PlexinD1 and Sema3E determine laminar positioning of heterotopically projecting callosal neurons. Mol. Cell. Neurosci. 2019, 100, 103397. [Google Scholar] [CrossRef]
- Burk, K.; Mire, E.; Bellon, A.; Hocine, M.; Guillot, J.; Moraes, F.; Yoshida, Y.; Simons, M.; Chauvet, S.; Mann, F. Post-endocytic sorting of Plexin-D1 controls signal transduction and development of axonal and vascular circuits. Nat. Commun. 2017, 8, 14508. [Google Scholar] [CrossRef] [Green Version]
- Mire, E.; Hocine, M.; Bazellières, E.; Jungas, T.; Davy, A.; Chauvet, S.; Mann, F. Developmental Upregulation of Ephrin-B1 Silences Sema3C/Neuropilin-1 Signaling during Post-crossing Navigation of Corpus Callosum Axons. Curr. Biol. 2018, 28, 1768–1782.e1764. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Bendito, G.; Flames, N.; Ma, L.; Fouquet, C.; Di Meglio, T.; Chedotal, A.; Tessier-Lavigne, M.; Marin, O. Robo1 and Robo2 cooperate to control the guidance of major axonal tracts in the mammalian forebrain. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 3395–3407. [Google Scholar] [CrossRef] [Green Version]
- Shu, T.; Sundaresan, V.; McCarthy, M.M.; Richards, L.J. Slit2 guides both precrossing and postcrossing callosal axons at the midline in vivo. J. Neurosci. Off. J. Soc. Neurosci. 2003, 23, 8176–8184. [Google Scholar] [CrossRef] [Green Version]
- Meijers, R.; Smock, R.G.; Zhang, Y.; Wang, J.H. Netrin Synergizes Signaling and Adhesion through DCC. Trends Biochem. Sci. 2020, 45, 6–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyer, N.P.; Gupton, S.L. Revisiting Netrin-1: One Who Guides (Axons). Front. Cell. Neurosci. 2018, 12, 221. [Google Scholar] [CrossRef] [PubMed]
- Fothergill, T.; Donahoo, A.L.; Douglass, A.; Zalucki, O.; Yuan, J.; Shu, T.; Goodhill, G.J.; Richards, L.J. Netrin-DCC signaling regulates corpus callosum formation through attraction of pioneering axons and by modulating Slit2-mediated repulsion. Cereb. Cortex 2014, 24, 1138–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charvet, C.J.; Das, A.; Song, J.W.; Tindal-Burgess, D.J.; Kabaria, P.; Dai, G.; Kane, T.; Takahashi, E. High Angular Resolution Diffusion MRI Reveals Conserved and Deviant Programs in the Paths that Guide Human Cortical Circuitry. Cereb Cortex 2020, 30, 1447–1464. [Google Scholar] [CrossRef]
- Manger, P.R.; Hemingway, J.; Haagensen, M.; Gilissen, E. Cross-sectional area of the elephant corpus callosum: Comparison to other eutherian mammals. Neuroscience 2010, 167, 815–824. [Google Scholar] [CrossRef]
- Hendrickson, T.J.; Mueller, B.A.; Sowell, E.R.; Mattson, S.N.; Coles, C.D.; Kable, J.A.; Jones, K.L.; Boys, C.J.; Lim, K.O.; Riley, E.P.; et al. Cortical gyrification is abnormal in children with prenatal alcohol exposure. Neuroimage. Clin. 2017, 15, 391–400. [Google Scholar] [CrossRef]
- Innocenti, G.M.; Price, D.J. Exuberance in the development of cortical networks. Nat. Rev. Neurosci. 2005, 6, 955–965. [Google Scholar] [CrossRef]
- Hand, R.A.; Khalid, S.; Tam, E.; Kolodkin, A.L. Axon Dynamics during Neocortical Laminar Innervation. Cell Rep. 2015, 12, 172–182. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, H.; Hirano, T.; Tagawa, Y. Pre-synaptic and post-synaptic neuronal activity supports the axon development of callosal projection neurons during different post-natal periods in the mouse cerebral cortex. Eur. J. Neurosci. 2010, 31, 410–424. [Google Scholar] [CrossRef]
- Fenlon, L.R.; Richards, L.J. Contralateral targeting of the corpus callosum in normal and pathological brain function. Trends Neurosci. 2015, 38, 264–272. [Google Scholar] [CrossRef]
- Wuttke, T.V.; Markopoulos, F.; Padmanabhan, H.; Wheeler, A.P.; Murthy, V.N.; Macklis, J.D. Developmentally primed cortical neurons maintain fidelity of differentiation and establish appropriate functional connectivity after transplantation. Nat. Neurosci. 2018, 21, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Rouaux, C.; Arlotta, P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat. Cell Biol. 2013, 15, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Leone, D.P.; Heavner, W.E.; Ferenczi, E.A.; Dobreva, G.; Huguenard, J.R.; Grosschedl, R.; McConnell, S.K. Satb2 Regulates the Differentiation of Both Callosal and Subcerebral Projection Neurons in the Developing Cerebral Cortex. Cereb Cortex 2015, 25, 3406–3419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Tornos, F.M.; Briz, C.G.; Weiss, L.A.; Sebastián-Serrano, A.; Ares, S.; Navarrete, M.; Frangeul, L.; Galazo, M.; Jabaudon, D.; Esteban, J.A.; et al. Cux1 Enables Interhemispheric Connections of Layer II/III Neurons by Regulating Kv1-Dependent Firing. Neuron 2016, 89, 494–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catapano, L.A.; Arnold, M.W.; Perez, F.A.; Macklis, J.D. Specific neurotrophic factors support the survival of cortical projection neurons at distinct stages of development. J. Neurosci. Off. J. Soc. Neurosci. 2001, 21, 8863–8872. [Google Scholar] [CrossRef] [Green Version]
- Ueno, M.; Fujita, Y.; Tanaka, T.; Nakamura, Y.; Kikuta, J.; Ishii, M.; Yamashita, T. Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci. 2013, 16, 543–551. [Google Scholar] [CrossRef]
- Poulopoulos, A.; Murphy, A.J.; Ozkan, A.; Davis, P.; Hatch, J.; Kirchner, R.; Macklis, J.D. Subcellular transcriptomes and proteomes of developing axon projections in the cerebral cortex. Nature 2019, 565, 356–360. [Google Scholar] [CrossRef]
- Tagawa, Y.; Hirano, T. Activity-dependent callosal axon projections in neonatal mouse cerebral cortex. Neural Plast 2012, 2012, 797295. [Google Scholar] [CrossRef] [Green Version]
- Ortiz, C.; Navarro, J.F.; Jurek, A.; Märtin, A.; Lundeberg, J.; Meletis, K. Molecular atlas of the adult mouse brain. Sci. Adv. 2020, 6, eabb3446. [Google Scholar] [CrossRef]
- Franco, S.J.; Gil-Sanz, C.; Martinez-Garay, I.; Espinosa, A.; Harkins-Perry, S.R.; Ramos, C.; Muller, U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science 2012, 337, 746–749. [Google Scholar] [CrossRef] [Green Version]
- Eckler, M.J.; Nguyen, T.D.; McKenna, W.L.; Fastow, B.L.; Guo, C.; Rubenstein, J.L.R.; Chen, B. Cux2-positive radial glial cells generate diverse subtypes of neocortical projection neurons and macroglia. Neuron 2015, 86, 1100–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Sanz, C.; Espinosa, A.; Fregoso, S.P.; Bluske, K.K.; Cunningham, C.L.; Martinez-Garay, I.; Zeng, H.; Franco, S.J.; Muller, U. Lineage Tracing Using Cux2-Cre and Cux2-CreERT2 Mice. Neuron 2015, 86, 1091–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, C.; Eckler, M.J.; McKenna, W.L.; McKinsey, G.L.; Rubenstein, J.L.; Chen, B. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron 2013, 80, 1167–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luarte, A.; Cornejo, V.H.; Bertin, F.; Gallardo, J.; Couve, A. The axonal endoplasmic reticulum: One organelle-many functions in development, maintenance, and plasticity. Dev. Neurobiol. 2018, 78, 181–208. [Google Scholar] [CrossRef] [PubMed]
- Silbereis, J.C.; Pochareddy, S.; Zhu, Y.; Li, M.; Sestan, N. The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 2016, 89, 248–268. [Google Scholar] [CrossRef] [Green Version]
- Sousa, A.M.M.; Zhu, Y.; Raghanti, M.A.; Kitchen, R.R.; Onorati, M.; Tebbenkamp, A.T.N.; Stutz, B.; Meyer, K.A.; Li, M.; Kawasawa, Y.I.; et al. Molecular and cellular reorganization of neural circuits in the human lineage. Science 2017, 358, 1027–1032. [Google Scholar] [CrossRef] [PubMed]


| Molecule | Molecular Function | Cortical Expression | CPN Subgroup Identification | Roles in CPN Development | References |
|---|---|---|---|---|---|
| CAV1 | Lipid-bound scaffolding domain protein | Layer V in caudo-lateral cortex, late embryonic to early postnatal period | Dual projecting callosal/frontal projection neurons (CPN/FPN) | Not necessary for early specification of CPN/FPN; not necessary for dual axonal targeting; may function in postmitotic development and refinement | [62,63] |
| LMO4 | Probable transcriptional factor | Layer V during early differentiation (E15.5), then expands to all cortical layer by P0 and later stages | CPNs and subcerebral projection neurons in presumptive sensory-motor area; colocalized with SATB2 in layer V by P6 | Second backward projection development; molecular identity diversification of CPNs in rostral motor cortex | [73,74,75] |
| CITED2 | Transcriptional coactivator of the p300/CBP-mediated transcription complex | Subventricular zone at E15.5; layer II/III, V, and VI in postnatal somatosensory cortex | CPNs in somatosensory cortex | Necessary for acquiring molecular identity of upper layer CPNs in somatosensory cortex | [11,63,76] |
| CTIP1 | DNA-binding transcription factor | Embryonic callosal and corticothalamic projection neurons; high in all layers of somatosensory cortex and the most superficial aspect of layer II/III in motor cortex postnatally | Expressed by all CPNs | Repression of CTIP2 expression; specification of sensory area identity in CPNs and other neurons | [77,78,79] |
| FEZF2 | DNA-binding transcription factor | Forebrain progenitors and their progeny in layer V | No | Repression of SATB2 expression; specification of subcerebral neuron identity | [48,60,80,81,82] |
| SNF2H | ATP-dependent chromatin remodeling protein | Embryonic neural progenitors | No | Primes upper layer cortical neuron development | [53,83,84] |
| INPP4B | Enzyme involved in phosphatidylinositol signaling pathways | TBD | No | Controlling axon polarization and generation of SATB2+ pyramidal neuron population | [54] |
| DOT1L | Histone methyltransferase specific to H3K79 | Progenitor zone and cortical plate | TBD | Regulation of SATB2 and CTIP2 expression | [51,52,85] |
| ASCL1/NGN2 | Basic helix-loop-helix family transcription factors | Neural progenitors in the embryonic ventral and dorsal telencephalon, respectively | No | Regulate the generation of SATB2+ upper layer neurons | [86,87,88] |
| FOXG1 | Forked-head family transcription factor | Neural progenitors in embryonic cortex; high in layer II/III and lower layer V postnatally | No | Promotes SATB2 expression and layer II/III CPN specification; directly represses Robo1 and Slit3 expression; directly represses Coup-TF1 expression | [66,68,89] |
| COUP-TF1 | Member of nuclear hormone receptor family of steroid hormone receptors | Superficial cortical plate in embryonic brain; layer IV and upper layer V postnatally | No | Promotes layer IV identity, while suppresses layer II/III and layer V specification | [68] |
| Molecule | Molecular Function | Cortical Expression | Interacting Pathway | Roles in Callosal Axon Development | Reference |
|---|---|---|---|---|---|
| APP | Receptor-like membrane protein | Embryonic and neonatal CC and neuronal cell body in layer V | Slit/Robo | Serves as a Slit receptor and mediates axon repulsion | [107] |
| HSC70 | Molecular chaperone of the heat shock protein 70 (HSP70) family | Preferentially expressed in neurons | Netrin/DCC | Required for the stability of DCC/TRIO complex at the growth cone to mediate axon outgrowth and guidance | [108] |
| PLCγ1 | Signal transducer of receptor tyrosine kinases | Broadly expressed in the brain from embryonic to adult stages; strongly expressed in the cortex | Netrin/DCC | Triggers actin rearrangement for axonal growth | [109] |
| MARCKS | Cellular substrate for protein kinase C; F-actin crosslinking protein | Ubiquitous | Netrin/DCC | Mediates DCC activation via membrane recruitment of tyrosine kinases PTK2 and SRC | [110,111] |
| GPM6A and GPM6B | Glycoprotein localized in cholesterol-rich lipid rafts of the plasma membrane | Expressed in actively elongating axons in embryonic and neonatal brain | Extension and guidance of callosal axons | [112,113] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ku, R.Y.; Torii, M. New Molecular Players in the Development of Callosal Projections. Cells 2021, 10, 29. https://doi.org/10.3390/cells10010029
Ku RY, Torii M. New Molecular Players in the Development of Callosal Projections. Cells. 2021; 10(1):29. https://doi.org/10.3390/cells10010029
Chicago/Turabian StyleKu, Ray Yueh, and Masaaki Torii. 2021. "New Molecular Players in the Development of Callosal Projections" Cells 10, no. 1: 29. https://doi.org/10.3390/cells10010029
APA StyleKu, R. Y., & Torii, M. (2021). New Molecular Players in the Development of Callosal Projections. Cells, 10(1), 29. https://doi.org/10.3390/cells10010029