RNA Profiling of Mouse Ependymal Cells after Spinal Cord Injury Identifies the Oncostatin Pathway as a Potential Key Regulator of Spinal Cord Stem Cell Fate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Cultures
2.3. QPCR
2.4. Western Blot
2.5. ELISA
2.6. Equipment and Settings
2.7. Spinal Cord Injury
2.8. Histology and Immunofluorescence
2.9. RNA Extraction, RNA Profilings and Bioinformatics Analysis
2.10. Statistical Analysis and Countings
3. Results
3.1. Ependymal Cell RNA Profile Is Highly Modified by SCI


3.2. Cilia Genes Are Downregulated after SCI and Are Controlled by Growth Factors
3.3. Oncostatin Affects Proliferation and Differentiation of Spinal Cord Neurosphere Cells
3.4. OSM Expression Is Strongly Upregulated in SCI
3.5. Microglia Upregulates Osmr in Spinal Cord Neurosphere Cells
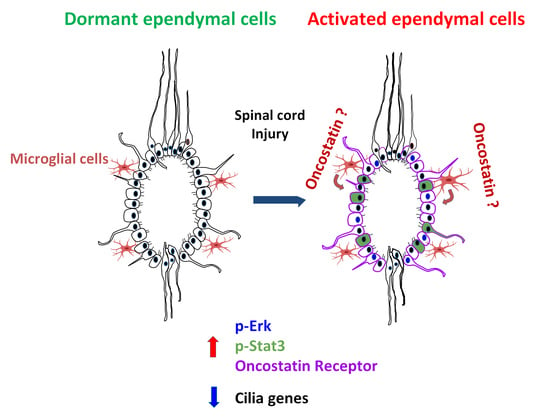
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Becker, C.G.; Becker, T.; Hugnot, J.-P. The Spinal Ependymal Zone as a Source of Endogenous Repair Cells across Vertebrates. Prog. Neurobiol. 2018, 170, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, L.K.; Truong, M.K.V.; Bednarczyk, M.R.; Aumont, A.; Fernandes, K.J.L. Cellular Organization of the Central Canal Ependymal Zone, a Niche of Latent Neural Stem Cells in the Adult Mammalian Spinal Cord. Neuroscience 2009, 164, 1044–1056. [Google Scholar] [CrossRef] [PubMed]
- Marichal, N.; Reali, C.; Trujillo-Cenóz, O.; Russo, R.E. Spinal Cord Stem Cells in Their Microenvironment: The Ependyma as a Stem Cell Niche. Adv. Exp. Med. Biol. 2017, 1041, 55–79. [Google Scholar] [CrossRef] [PubMed]
- Sabelström, H.; Stenudd, M.; Frisén, J. Neural Stem Cells in the Adult Spinal Cord. Exp. Neurol. 2014, 260, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ovejero, D.; Arevalo-Martin, A.; Paniagua-Torija, B.; Florensa-Vila, J.; Ferrer, I.; Grassner, L.; Molina-Holgado, E. The Ependymal Region of the Adult Human Spinal Cord Differs from Other Species and Shows Ependymoma-like Features. Brain J. Neurol. 2015, 138, 1583–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghazale, H.; Ripoll, C.; Leventoux, N.; Jacob, L.; Azar, S.; Mamaeva, D.; Glasson, Y.; Calvo, C.-F.; Thomas, J.-L.; Meneceur, S.; et al. RNA Profiling of the Human and Mouse Spinal Cord Stem Cell Niches Reveals an Embryonic-like Regionalization with MSX1+ Roof-Plate-Derived Cells. Stem Cell Rep. 2019, 12, 1159–1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfaro-Cervello, C.; Soriano-Navarro, M.; Mirzadeh, Z.; Alvarez-Buylla, A.; Garcia-Verdugo, J.M. Biciliated Ependymal Cell Proliferation Contributes to Spinal Cord Growth. J. Comp. Neurol. 2012, 520, 3528–3552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfenninger, C.V.; Steinhoff, C.; Hertwig, F.; Nuber, U.A. Prospectively Isolated CD133/CD24-Positive Ependymal Cells from the Adult Spinal Cord and Lateral Ventricle Wall Differ in Their Long-Term in Vitro Self-Renewal and in Vivo Gene Expression. Glia 2011, 59, 68–81. [Google Scholar] [CrossRef]
- MacDonald, A.; Lu, B.; Caron, M.; Caporicci-Dinucci, N.; Hatrock, D.; Petrecca, K.; Bourque, G.; Stratton, J.A. Single Cell Transcriptomics of Ependymal Cells Across Age, Region and Species Reveals Cilia-Related and Metal Ion Regulatory Roles as Major Conserved Ependymal Cell Functions. Front. Cell. Neurosci. 2021, 15, 268. [Google Scholar] [CrossRef]
- Llorens-Bobadilla, E.; Chell, J.M.; Le Merre, P.; Wu, Y.; Zamboni, M.; Bergenstråhle, J.; Stenudd, M.; Sopova, E.; Lundeberg, J.; Shupliakov, O.; et al. A Latent Lineage Potential in Resident Neural Stem Cells Enables Spinal Cord Repair. Science 2020, 370, eabb8795. [Google Scholar] [CrossRef]
- Adrian, E.K.; Walker, B.E. Incorporation of Thymidine-H3 by Cells in Normal and Injured Mouse Spinal Cord. J. Neuropathol. Exp. Neurol. 1962, 21, 597–609. [Google Scholar] [CrossRef] [Green Version]
- Sabelström, H.; Stenudd, M.; Réu, P.; Dias, D.O.; Elfineh, M.; Zdunek, S.; Damberg, P.; Göritz, C.; Frisén, J. Resident Neural Stem Cells Restrict Tissue Damage and Neuronal Loss After Spinal Cord Injury in Mice. Science 2013, 342, 637–640. [Google Scholar] [CrossRef] [Green Version]
- Ren, Y.; Ao, Y.; O’Shea, T.M.; Burda, J.E.; Bernstein, A.M.; Brumm, A.J.; Muthusamy, N.; Ghashghaei, H.T.; Carmichael, S.T.; Cheng, L.; et al. Ependymal Cell Contribution to Scar Formation after Spinal Cord Injury Is Minimal, Local and Dependent on Direct Ependymal Injury. Sci. Rep. 2017, 7, 41122. [Google Scholar] [CrossRef] [Green Version]
- Stenudd, M.; Sabelström, H.; Frisén, J. Role of Endogenous Neural Stem Cells in Spinal Cord Injury and Repair. JAMA Neurol. 2015, 72, 235–237. [Google Scholar] [CrossRef]
- Meletis, K.; Barnabé-Heider, F.; Carlén, M.; Evergren, E.; Tomilin, N.; Shupliakov, O.; Frisén, J. Spinal Cord Injury Reveals Multilineage Differentiation of Ependymal Cells. PLoS Biol. 2008, 6, e182. [Google Scholar] [CrossRef]
- Barnabé-Heider, F.; Göritz, C.; Sabelström, H.; Takebayashi, H.; Pfrieger, F.W.; Meletis, K.; Frisén, J. Origin of New Glial Cells in Intact and Injured Adult Spinal Cord. Cell Stem Cell 2010, 7, 470–482. [Google Scholar] [CrossRef] [Green Version]
- Shihabuddin, L.S.; Horner, P.J.; Ray, J.; Gage, F.H. Adult Spinal Cord Stem Cells Generate Neurons after Transplantation in the Adult Dentate Gyrus. J. Neurosci. Off. J. Soc. Neurosci. 2000, 20, 8727–8735. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.; Aliberti, J.; Graemmel, P.; Sunshine, M.J.; Kreutzberg, G.W.; Sher, A.; Littman, D.R. Analysis of Fractalkine Receptor CX(3)CR1 Function by Targeted Deletion and Green Fluorescent Protein Reporter Gene Insertion. Mol. Cell. Biol. 2000, 20, 4106–4114. [Google Scholar] [CrossRef] [Green Version]
- Okabe, M.; Ikawa, M.; Kominami, K.; Nakanishi, T.; Nishimune, Y. “Green Mice” as a Source of Ubiquitous Green Cells. FEBS Lett. 1997, 407, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Hugnot, J.-P. Isolate and Culture Neural Stem Cells from the Mouse Adult Spinal Cord. Methods Mol. Biol. Clifton NJ 2022, 2389, 45–56. [Google Scholar] [CrossRef]
- Blasi, E.; Barluzzi, R.; Bocchini, V.; Mazzolla, R.; Bistoni, F. Immortalization of Murine Microglial Cells by a V-Raf/v-Myc Carrying Retrovirus. J. Neuroimmunol. 1990, 27, 229–237. [Google Scholar] [CrossRef]
- Fernandez-Zafra, T.; Codeluppi, S.; Uhlén, P. An Ex Vivo Spinal Cord Injury Model to Study Ependymal Cells in Adult Mouse Tissue. Exp. Cell Res. 2017, 357, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Azar, S.; Leventoux, N.; Ripoll, C.; Rigau, V.; Gozé, C.; Lorcy, F.; Bauchet, L.; Duffau, H.; Guichet, P.O.; Rothhut, B.; et al. Cellular and Molecular Characterization of IDH1-Mutated Diffuse Low Grade Gliomas Reveals Tumor Heterogeneity and Absence of EGFR/PDGFRα Activation. Glia 2018, 66, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Lockard, I.; Reers, B.L. Staining Tissue of the Central Nervous System with Luxol Fast Blue and Neutral Red. Stain Technol. 1962, 37, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinforma. Oxf. Engl. 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Clèries, R.; Galvez, J.; Espino, M.; Ribes, J.; Nunes, V.; de Heredia, M.L. BootstRatio: A Web-Based Statistical Analysis of Fold-Change in QPCR and RT-QPCR Data Using Resampling Methods. Comput. Biol. Med. 2012, 42, 438–445. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, Y.; Vogel, L.C.; DeVivo, M.J. Causes of Spinal Cord Injury. Top. Spinal Cord Inj. Rehabil. 2013, 19, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Velmahos, G.C.; Degiannis, E.; Hart, K.; Souter, I.; Saadia, R. Changing Profiles in Spinal Cord Injuries and Risk Factors Influencing Recovery after Penetrating Injuries. J. Trauma 1995, 38, 334–337. [Google Scholar] [CrossRef]
- Hallen, A.; Cooper, A.J.L.; Jamie, J.F.; Karuso, P. Insights into Enzyme Catalysis and Thyroid Hormone Regulation of Cerebral Ketimine Reductase/μ-Crystallin Under Physiological Conditions. Neurochem. Res. 2015, 40, 1252–1266. [Google Scholar] [CrossRef]
- Siegenthaler, G. Extra-and Intracellular Transport of Retinoids: A Reappraisal. Horm. Res. 1996, 45, 122–127. [Google Scholar] [CrossRef]
- Clark, D.W.; Mitra, A.; Fillmore, R.A.; Jiang, W.G.; Samant, R.S.; Fodstad, O.; Shevde, L.A. NUPR1 Interacts with P53, Transcriptionally Regulates P21 and Rescues Breast Epithelial Cells from Doxorubicin-Induced Genotoxic Stress. Curr. Cancer Drug Targets 2008, 8, 421–430. [Google Scholar] [CrossRef]
- Xin, H.; D’Souza, S.; Jørgensen, T.N.; Vaughan, A.T.; Lengyel, P.; Kotzin, B.L.; Choubey, D. Increased Expression of Ifi202, an IFN-Activatable Gene, in B6.Nba2 Lupus Susceptible Mice Inhibits P53-Mediated Apoptosis. J. Immunol. Baltim. Md 1950 2006, 176, 5863–5870. [Google Scholar] [CrossRef] [Green Version]
- Sercu, S.; Zhang, L.; Merregaert, J. The Extracellular Matrix Protein 1: Its Molecular Interaction and Implication in Tumor Progression. Cancer Investig. 2008, 26, 375–384. [Google Scholar] [CrossRef]
- Morikawa, Y. Oncostatin M in the Development of the Nervous System. Anat. Sci. Int. 2005, 80, 53–59. [Google Scholar] [CrossRef]
- Balamurugan, K.; Sterneck, E. The Many Faces of C/EBPδ and Their Relevance for Inflammation and Cancer. Int. J. Biol. Sci. 2013, 9, 917–933. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Newbern, J.; Wu, Y.; Morgan-Smith, M.; Zhong, J.; Charron, J.; Snider, W. MEK Is a Key Regulator of Gliogenesis in the Developing Brain. Neuron 2012, 75, 1035–1050. [Google Scholar] [CrossRef] [Green Version]
- Kovács, K.J. C-Fos as a Transcription Factor: A Stressful (Re)View from a Functional Map. Neurochem. Int. 1998, 33, 287–297. [Google Scholar] [CrossRef]
- Sun, K.L.W.; Correia, J.P.; Kennedy, T.E. Netrins: Versatile Extracellular Cues with Diverse Functions. Dev. Camb. Engl. 2011, 138, 2153–2169. [Google Scholar] [CrossRef] [Green Version]
- Goto, H.; Inaba, H.; Inagaki, M. Mechanisms of Ciliogenesis Suppression in Dividing Cells. Cell. Mol. Life Sci. CMLS 2017, 74, 881–890. [Google Scholar] [CrossRef] [Green Version]
- Goto, H.; Inoko, A.; Inagaki, M. Cell Cycle Progression by the Repression of Primary Cilia Formation in Proliferating Cells. Cell. Mol. Life Sci. CMLS 2013, 70, 3893–3905. [Google Scholar] [CrossRef] [Green Version]
- Goranci-Buzhala, G.; Mariappan, A.; Ricci-Vitiani, L.; Josipovic, N.; Pacioni, S.; Gottardo, M.; Ptok, J.; Schaal, H.; Callaini, G.; Rajalingam, K.; et al. Cilium Induction Triggers Differentiation of Glioma Stem Cells. Cell Rep. 2021, 36, 109656. [Google Scholar] [CrossRef]
- Lemeille, S.; Paschaki, M.; Baas, D.; Morlé, L.; Duteyrat, J.-L.; Ait-Lounis, A.; Barras, E.; Soulavie, F.; Jerber, J.; Thomas, J.; et al. Interplay of RFX Transcription Factors 1, 2 and 3 in Motile Ciliogenesis. Nucleic Acids Res. 2020, 48, 9019–9036. [Google Scholar] [CrossRef]
- Nemajerova, A.; Moll, U.M. Tissue-Specific Roles of P73 in Development and Homeostasis. J. Cell Sci. 2019, 132, jcs233338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, M.; Stracker, T.H. Transcriptional Regulation of Multiciliated Cell Differentiation. Semin. Cell Dev. Biol. 2020, 110, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Abdi, K.; Lai, C.-H.; Paez-Gonzalez, P.; Lay, M.; Pyun, J.; Kuo, C.T. Uncovering Inherent Cellular Plasticity of Multiciliated Ependyma Leading to Ventricular Wall Transformation and Hydrocephalus. Nat. Commun. 2018, 9, 1655. [Google Scholar] [CrossRef] [PubMed]
- Sabourin, J.-C.; Ackema, K.B.; Ohayon, D.; Guichet, P.-O.; Perrin, F.E.; Garces, A.; Ripoll, C.; Charité, J.; Simonneau, L.; Kettenmann, H.; et al. A Mesenchymal-like ZEB1(+) Niche Harbors Dorsal Radial Glial Fibrillary Acidic Protein-Positive Stem Cells in the Spinal Cord. Stem Cells Dayt. Ohio 2009, 27, 2722–2733. [Google Scholar] [CrossRef]
- Taga, T.; Fukuda, S. Role of IL-6 in the Neural Stem Cell Differentiation. Clin. Rev. Allergy Immunol. 2005, 28, 249–256. [Google Scholar] [CrossRef]
- Beatus, P.; Jhaveri, D.J.; Walker, T.L.; Lucas, P.G.; Rietze, R.L.; Cooper, H.M.; Morikawa, Y.; Bartlett, P.F. Oncostatin M Regulates Neural Precursor Activity in the Adult Brain. Dev. Neurobiol. 2011, 71, 619–633. [Google Scholar] [CrossRef]
- Tamura, S.; Morikawa, Y.; Senba, E. Localization of Oncostatin M Receptor Beta in Adult and Developing CNS. Neuroscience 2003, 119, 991–997. [Google Scholar] [CrossRef]
- Chen, K.; Deng, S.; Lu, H.; Zheng, Y.; Yang, G.; Kim, D.; Cao, Q.; Wu, J.Q. RNA-Seq Characterization of Spinal Cord Injury Transcriptome in Acute/Subacute Phases: A Resource for Understanding the Pathology at the Systems Level. PLoS ONE 2013, 8, e72567. [Google Scholar] [CrossRef] [Green Version]
- Milich, L.M.; Choi, J.S.; Ryan, C.; Cerqueira, S.R.; Benavides, S.; Yahn, S.L.; Tsoulfas, P.; Lee, J.K. Single-Cell Analysis of the Cellular Heterogeneity and Interactions in the Injured Mouse Spinal Cord. J. Exp. Med. 2021, 218, e20210040. [Google Scholar] [CrossRef]
- Matarredona, E.R.; Talaverón, R.; Pastor, A.M. Interactions Between Neural Progenitor Cells and Microglia in the Subventricular Zone: Physiological Implications in the Neurogenic Niche and After Implantation in the Injured Brain. Front. Cell. Neurosci. 2018, 12, 268. [Google Scholar] [CrossRef]
- Heintz, N. Gene Expression Nervous System Atlas (GENSAT). Nat. Neurosci. 2004, 7, 483. [Google Scholar] [CrossRef]
- He, Y.; Yao, X.; Taylor, N.; Bai, Y.; Lovenberg, T.; Bhattacharya, A. RNA Sequencing Analysis Reveals Quiescent Microglia Isolation Methods from Postnatal Mouse Brains and Limitations of BV2 Cells. J. Neuroinflam. 2018, 15, 153. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a Unique TGF-β Dependent Molecular and Functional Signature in Microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Henn, A.; Lund, S.; Hedtjärn, M.; Schrattenholz, A.; Pörzgen, P.; Leist, M. The Suitability of BV2 Cells as Alternative Model System for Primary Microglia Cultures or for Animal Experiments Examining Brain Inflammation. ALTEX 2009, 26, 83–94. [Google Scholar] [CrossRef] [Green Version]
- Timmerman, R.; Burm, S.M.; Bajramovic, J.J. An Overview of in Vitro Methods to Study Microglia. Front. Cell. Neurosci. 2018, 12, 242. [Google Scholar] [CrossRef]
- Balanis, N.; Wendt, M.K.; Schiemann, B.J.; Wang, Z.; Schiemann, W.P.; Carlin, C.R. Epithelial to Mesenchymal Transition Promotes Breast Cancer Progression via a Fibronectin-Dependent STAT3 Signaling Pathway. J. Biol. Chem. 2013, 288, 17954–17967. [Google Scholar] [CrossRef] [Green Version]
- Gu, F.; Hata, R.; Ma, Y.-J.; Tanaka, J.; Mitsuda, N.; Kumon, Y.; Hanakawa, Y.; Hashimoto, K.; Nakajima, K.; Sakanaka, M. Suppression of Stat3 Promotes Neurogenesis in Cultured Neural Stem Cells. J. Neurosci. Res. 2005, 81, 163–171. [Google Scholar] [CrossRef]
- Natarajan, R.; Singal, V.; Benes, R.; Gao, J.; Chan, H.; Chen, H.; Yu, Y.; Zhou, J.; Wu, P. STAT3 Modulation to Enhance Motor Neuron Differentiation in Human Neural Stem Cells. PLoS ONE 2014, 9, e100405. [Google Scholar] [CrossRef]
- Li, T.; Zhao, X.; Duan, J.; Cui, S.; Zhu, K.; Wan, Y.; Liu, S.; Peng, Z.; Wang, L. Targeted Inhibition of STAT3 in Neural Stem Cells Promotes Neuronal Differentiation and Functional Recovery in Rats with Spinal Cord Injury. Exp. Ther. Med. 2021, 22, 711. [Google Scholar] [CrossRef]
- Niwa, H.; Burdon, T.; Chambers, I.; Smith, A. Self-Renewal of Pluripotent Embryonic Stem Cells Is Mediated via Activation of STAT. Genes Dev. 1998, 12, 2048–2060. [Google Scholar] [CrossRef] [Green Version]
- Heon Rhim, J.; Luo, X.; Gao, D.; Xu, X.; Zhou, T.; Li, F.; Wang, P.; Wong, S.T.C.; Xia, X. Cell Type-Dependent Erk-Akt Pathway Crosstalk Regulates the Proliferation of Fetal Neural Progenitor Cells. Sci. Rep. 2016, 6, 26547. [Google Scholar] [CrossRef]
- He, L.; Gu, W.; Wang, M.; Chang, X.; Sun, X.; Zhang, Y.; Lin, X.; Yan, C.; Fan, W.; Su, P.; et al. Extracellular Matrix Protein 1 Promotes Follicular Helper T Cell Differentiation and Antibody Production. Proc. Natl. Acad. Sci. USA 2018, 115, 8621–8626. [Google Scholar] [CrossRef] [Green Version]
- Domenzain-Reyna, C.; Hernández, D.; Miquel-Serra, L.; Docampo, M.J.; Badenas, C.; Fabra, A.; Bassols, A. Structure and Regulation of the Versican Promoter: The Versican Promoter Is Regulated by AP-1 and TCF Transcription Factors in Invasive Human Melanoma Cells. J. Biol. Chem. 2009, 284, 12306–12317. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Pan, J.; Dong, Y.; Tweardy, D.J.; Dong, Y.; Garibotto, G.; Mitch, W.E. Stat3 Activation Links a C/EBPδ to Myostatin Pathway to Stimulate Loss of Muscle Mass. Cell Metab. 2013, 18, 368–379. [Google Scholar] [CrossRef] [Green Version]
- Higgins, M.; Obaidi, I.; McMorrow, T. Primary Cilia and Their Role in Cancer. Oncol. Lett. 2019, 17, 3041–3047. [Google Scholar] [CrossRef]
- Yanagisawa, M.; Nakashima, K.; Taga, T. STAT3-Mediated Astrocyte Differentiation from Mouse Fetal Neuroepithelial Cells by Mouse Oncostatin M. Neurosci. Lett. 1999, 269, 169–172. [Google Scholar] [CrossRef]
- Stross, C.; Radtke, S.; Clahsen, T.; Gerlach, C.; Volkmer-Engert, R.; Schaper, F.; Heinrich, P.C.; Hermanns, H.M. Oncostatin M Receptor-Mediated Signal Transduction Is Negatively Regulated by SOCS3 through a Receptor Tyrosine-Independent Mechanism. J. Biol. Chem. 2006, 281, 8458–8468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliva, A.A.; Kang, Y.; Sanchez-Molano, J.; Furones, C.; Atkins, C.M. STAT3 Signaling after Traumatic Brain Injury. J. Neurochem. 2012, 120, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Slaets, H.; Nelissen, S.; Janssens, K.; Vidal, P.M.; Lemmens, E.; Stinissen, P.; Hendrix, S.; Hellings, N. Oncostatin M Reduces Lesion Size and Promotes Functional Recovery and Neurite Outgrowth after Spinal Cord Injury. Mol. Neurobiol. 2014, 50, 1142–1151. [Google Scholar] [CrossRef]
- Nakanishi, M.; Niidome, T.; Matsuda, S.; Akaike, A.; Kihara, T.; Sugimoto, H. Microglia-Derived Interleukin-6 and Leukaemia Inhibitory Factor Promote Astrocytic Differentiation of Neural Stem/Progenitor Cells. Eur. J. Neurosci. 2007, 25, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Hata, R.; Cao, F.; Gu, F.; Hanakawa, Y.; Hashimoto, K.; Sakanaka, M. Ramified Microglial Cells Promote Astrogliogenesis and Maintenance of Neural Stem Cells through Activation of Stat3 Function. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008, 22, 3866–3877. [Google Scholar] [CrossRef] [Green Version]
- Banner, L.R.; Moayeri, N.N.; Patterson, P.H. Leukemia Inhibitory Factor Is Expressed in Astrocytes Following Cortical Brain Injury. Exp. Neurol. 1997, 147, 1–9. [Google Scholar] [CrossRef]
- Ganesh, K.; Das, A.; Dickerson, R.; Khanna, S.; Parinandi, N.L.; Gordillo, G.M.; Sen, C.K.; Roy, S. Prostaglandin E2 Induces Oncostatin M Expression in Human Chronic Wound Macrophages through Axl Receptor Tyrosine Kinase Pathway. J. Immunol. 2012, 189, 2563–2573. [Google Scholar] [CrossRef] [Green Version]
- Lakshman, N.; Bourget, C.; Siu, R.; Bamm, V.V.; Xu, W.; Harauz, G.; Morshead, C.M. Niche-Dependent Inhibition of Neural Stem Cell Proliferation and Oligodendrogenesis Is Mediated by the Presence of Myelin Basic Protein. Stem Cells Dayt. Ohio 2021, 39, 776–786. [Google Scholar] [CrossRef]
- Kyritsis, N.; Kizil, C.; Brand, M. Neuroinflammation and Central Nervous System Regeneration in Vertebrates. Trends Cell Biol. 2014, 24, 128–135. [Google Scholar] [CrossRef]




| Fold Change > 10 | ||||
| Gene Symbol | Description | Fold Change | p-Value | FDR |
| Crym | crystallin. mu | 33.3 | 1.6 × 10−8 | 2.5 × 10−5 |
| Osmr | oncostatin M receptor | 30.3 | 2.6 × 10−10 | 1.7 × 10−6 |
| Rbp1 | retinol binding protein 1. cellular | 28.8 | 1.8 × 10−7 | 0.0001 |
| Nupr1 | nuclear protein transcription regulator 1 | 26 | 2.2 × 10−6 | 0.0006 |
| Thbs2 | thrombospondin 2 | 25.8 | 3.8 × 10−12 | 5.6 × 10−8 |
| Ecm1 | extracellular matrix protein 1 | 24.1 | 4.5 × 10−10 | 2.2 × 10−6 |
| Ifi202b | interferon activated gene 202B; | 22.8 | 2.6 × 10−7 | 0.0001 |
| Cebpd | CCAAT/enhancer binding protein (C/EBP). delta | 15.4 | 1.0 × 10−5 | 0.0017 |
| Timp1 | tissue inhibitor of metalloproteinase 1 | 15.3 | 8.1 × 10−10 | 2.7 × 10−6 |
| Socs3 | suppressor of cytokine signaling 3 | 14.4 | 5.1 × 10−8 | 5.2 × 10−5 |
| Olfml3 | olfactomedin-like 3 | 13.3 | 1.7 × 10−10 | 1.3 × 10−6 |
| Gfap | glial fibrillary acidic protein | 12.9 | 9.8 × 10−7 | 0.0004 |
| Fos | FBJ osteosarcoma oncogene | 12.7 | 1.1 × 10−7 | 8.4 × 10−5 |
| Etv5 | ets variant 5 | 12.6 | 3.1 × 10−10 | 1.8 × 10−6 |
| Serping1 | serine peptidase inhibitor. clade G. member 1 | 12.2 | 9.6 × 10−9 | 1.8 × 10−5 |
| Ptx3 | pentraxin related gene | 12.2 | 9.4 × 10−7 | 0.0004 |
| Fgfrl1 | fibroblast growth factor receptor-like 1 | 11.9 | 5.5 × 10−8 | 5.5 × 10−5 |
| Vcan | versican | 11.6 | 2.9 × 10−5 | 0.003 |
| S100a6 | S100 calcium binding protein A6 (calcyclin) | 11.3 | 0.0006 | 0.0265 |
| Serpina3n | serine peptidase inhibitor. clade A. member 3N | 11.3 | 4.2 × 10−9 | 1.1 × 10−5 |
| Gm42151 | predicted gene. 42151 | 11.1 | 8.2 × 10−10 | 2.7 × 10−6 |
| Lgals1 | lectin. galactose binding. soluble 1 | 11.1 | 2.0 × 10−6 | 0.0006 |
| Fgl2 | fibrinogen-like protein 2 | 10.7 | 1.4 × 10−8 | 2.2 10−5 |
| Gbp3 | guanylate binding protein 3 | 10.1 | 0.017 | 0.23 |
| Fold Change < −4 | ||||
| Gene Symbol | Description | Fold Change | p-Value | FDR |
| Acta1 | actin. alpha 1. skeletal muscle | −21.2 | 0.035 | 0.35 |
| Pcsk2 | proprotein convertase subtilisin/kexin type 2 | −7.4 | 1.3 × 10−6 | 0.0004 |
| Snca | synuclein. alpha | −5.8 | 0.0009 | 0.036 |
| Myh1 | myosin. heavy polypeptide 1. skeletal muscle. adult | −5.3 | 0.007 | 0.13 |
| Snca | synuclein. alpha | −5.2 | 0.018 | 0.24 |
| Tchh | trichohyalin | −4.7 | 0.0005 | 0.027 |
| Myh1 | myosin. heavy polypeptide 1. skeletal muscle. adult | −4.7 | 0.007 | 0.137 |
| Chd3os | chromodomain helicase DNA binding protein 3. opposite strand | −4.6 | 1.5 × 10−5 | 0.0021 |
| Mb | myoglobin | −4.6 | 0.0001 | 0.008 |
| Sema5a | Semaphorin 5A | −4.4 | 0.015 | 0.22 |
| Snhg11 | small nucleolar RNA host gene 11 | −4.4 | 0.006 | 0.11 |
| Ccdc108 | coiled-coil domain containing 108 | −4.2 | 3.5 × 10−6 | 0.0008 |
| Tnni2 | troponin I. skeletal. fast 2 | −4.1 | 0.02 | 0.254 |
| Plppr4 | phospholipid phosphatase related 4 | −4.1 | 3.3 × 10−5 | 0.004 |
| Clstn2 | calsyntenin 2 | −4.1 | 0.002 | 0.05 |
| Tnni3 | troponin I. cardiac 3 | −4.0 | 6.6 × 10−5 | 0.006 |
| Acsl3 | acyl-CoA synthetase long-chain family member 3 | −4.0 | 0.019 | 0.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chevreau, R.; Ghazale, H.; Ripoll, C.; Chalfouh, C.; Delarue, Q.; Hemonnot-Girard, A.L.; Mamaeva, D.; Hirbec, H.; Rothhut, B.; Wahane, S.; et al. RNA Profiling of Mouse Ependymal Cells after Spinal Cord Injury Identifies the Oncostatin Pathway as a Potential Key Regulator of Spinal Cord Stem Cell Fate. Cells 2021, 10, 3332. https://doi.org/10.3390/cells10123332
Chevreau R, Ghazale H, Ripoll C, Chalfouh C, Delarue Q, Hemonnot-Girard AL, Mamaeva D, Hirbec H, Rothhut B, Wahane S, et al. RNA Profiling of Mouse Ependymal Cells after Spinal Cord Injury Identifies the Oncostatin Pathway as a Potential Key Regulator of Spinal Cord Stem Cell Fate. Cells. 2021; 10(12):3332. https://doi.org/10.3390/cells10123332
Chicago/Turabian StyleChevreau, Robert, Hussein Ghazale, Chantal Ripoll, Chaima Chalfouh, Quentin Delarue, Anne Laure Hemonnot-Girard, Daria Mamaeva, Helene Hirbec, Bernard Rothhut, Shalaka Wahane, and et al. 2021. "RNA Profiling of Mouse Ependymal Cells after Spinal Cord Injury Identifies the Oncostatin Pathway as a Potential Key Regulator of Spinal Cord Stem Cell Fate" Cells 10, no. 12: 3332. https://doi.org/10.3390/cells10123332
APA StyleChevreau, R., Ghazale, H., Ripoll, C., Chalfouh, C., Delarue, Q., Hemonnot-Girard, A. L., Mamaeva, D., Hirbec, H., Rothhut, B., Wahane, S., Perrin, F. E., Noristani, H. N., Guerout, N., & Hugnot, J. P. (2021). RNA Profiling of Mouse Ependymal Cells after Spinal Cord Injury Identifies the Oncostatin Pathway as a Potential Key Regulator of Spinal Cord Stem Cell Fate. Cells, 10(12), 3332. https://doi.org/10.3390/cells10123332