Human Genetic Variation Influences Enteric Fever Progression
Abstract
:1. Introduction
2. Host Genetic Variants
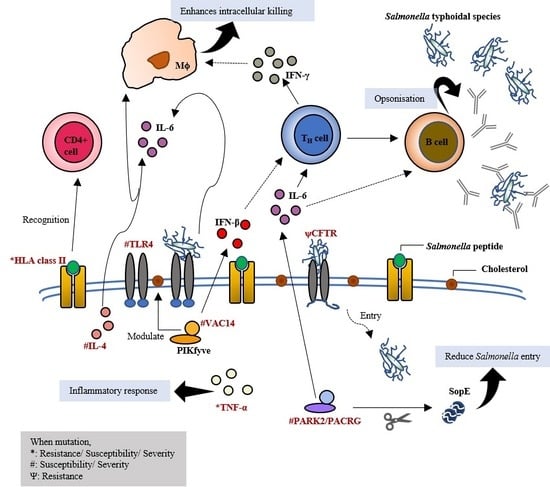
2.1. Toll-Like Receptor 4: Initiates the First Line of Defense
2.2. Toll-Like Receptor 5: Recognises Bacterial Flagellin
2.3. Natural Resistance-Associated Macrophage Protein 1: Kills Intracellular Pathogens
2.4. VAC14: Acts as Core Subunit of Lipid Kinase in Signalling the Type I IFNs Production
2.5. Variable Number of Tandem Repeat (VNTR) in Interleukin 4: Generating Variation in Gene Expression
2.6. PARK2/PACRG: Ubiquitination
2.7. Cystic Fibrosis Transmembrane Conductance Regulator: Secretion Channel and Pili Receptor
2.8. MHC Class II and Class III: Antigen Presentation and Other Innate Immunity Responses
3. Limitations
3.1. Samples Size and Population
3.2. Other Biological Specimens
3.3. Other Approaches
4. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wain, J.; Hendriksen, R.S.; Mikoleit, M.; Keddy, K.H.; Ochiai, R.L. Typhoid fever. Lancet 2014, 385, 1136–1145. [Google Scholar] [CrossRef]
- Crump, J.A. Progress in Typhoid Fever Epidemiology. Clin. Infect. Dis. 2019, 68, S4–S9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchello, C.S.; Hong, C.Y.; Crump, J.A. Global Typhoid Fever Incidence: A Systematic Review and Meta-analysis. Clin. Infect. Dis. 2019, 68, S105–S116. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Cortes, A.; Bessudo, D.; Sanchez-Leyva, R.; Fragoso, R.; Hinojosa, M.; Becerril, P. Water-borne transmission of chloramphenicol-resistant Salmonella typhi in Mexico. Lancet 1973, 302, 605–607. [Google Scholar] [CrossRef]
- Baker, S.; Karkey, A.; Parry, C. Are we adequately prepared for the emergence of Salmonella enterica serovar Paratyphi A? Lancet Glob. Health 2014, 2, e195–e196. [Google Scholar] [CrossRef] [Green Version]
- Antillón, M.; Warren, J.L.; Crawford, F.W.; Weinberger, D.M.; Kürüm, E.; Pak, G.D.; Marks, F.; Pitzer, V.E. The burden of typhoid fever in low- and middle-income countries: A meta-regression approach. PLoS Neglected Trop. Dis. 2017, 11, e0005376. [Google Scholar] [CrossRef]
- Mogasale, V.; Maskery, B.; Ochiai, R.L.; Lee, J.S.; Mogasale, V.V.; Ramani, E.; Kim, Y.E.; Park, J.K.; Wierzba, T.F. Burden of typhoid fever in low-income and middle-income countries: A systematic, literature-based update with risk-factor adjustment. Lancet Glob. Health 2014, 2, e570–e580. [Google Scholar] [CrossRef] [Green Version]
- Chau, T.T.; Campbell, J.I.; Galindo, C.M.; Hoang, N.V.M.; Diep, T.S.; Nga, T.T.T.; Chau, N.V.V.; Tuan, P.Q.; Page, A.L.; Ochiai, R.L.; et al. Antimicrobial Drug Resistance of Salmonella enterica Serovar Typhi in Asia and Molecular Mechanism of Reduced Susceptibility to the Fluoroquinolones. Antimicrob. Agents Chemother. 2007, 51, 4315–4323. [Google Scholar] [CrossRef] [Green Version]
- Naghavi, M.; Abajobir, A.A.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abera, S.F.; Aboyans, V.; Adetokunboh, O.; Afshin, A.; Agrawal, A.; et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef] [Green Version]
- Marks, F.; Von Kalckreuth, V.; Aaby, P.; Adu-Sarkodie, Y.; El Tayeb, M.A.; Ali, M.; Aseffa, A.; Baker, S.; Biggs, H.M.; Bjerregaard-Andersen, M.; et al. Incidence of invasive salmonella disease in sub-Saharan Africa: A multicentre population-based surveillance study. Lancet Glob. Health 2017, 5, e310–e323. [Google Scholar] [CrossRef] [Green Version]
- Majumder, P.P. Genomics of immune response to typhoid and cholera vaccines. Philos. Trans. R. Soc. B: Biol. Sci. 2015, 370, 20140142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, S.; Vollaard, A.M.; Widjaja, S.; Surjadi, C.; Van De Vosse, E.; Van Dissel, J.T. PARK2/PACRG polymorphisms and susceptibility to typhoid and paratyphoid fever. Clin. Exp. Immunol. 2006, 144, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Bhuvanendran, S.; Hussin, H.M.; Meran, L.P.; Anthony, A.A.; Zhang, L.; Burch, L.H.; Phua, K.K.; Ismail, A.; Balaram, P. Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms and typhoid susceptibility in Asian Malay population in Malaysia. Microbes Infect. 2011, 13, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Ziakas, P.D.; Prodromou, M.L.; El Khoury, J.; Zintzaras, E.; Mylonakis, E. The Role of TLR4 896 A>G and 1196 C>T in Susceptibility to Infections: A Review and Meta-Analysis of Genetic Association Studies. PLoS ONE 2013, 8, e81047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadl, M.A.; Aydarous, M.A.; Mao, C.; Yasmeen, A. An association of VNTR polymorphism in intron3 of IL-4 gene with susceptibility to typhoid fever in Khartoum State, Sudan. Kuwait J. Sci. 2016, 43, 185–192. [Google Scholar]
- Alvarez, M.I.; Glover, L.C.; Luo, P.; Wang, L.; Theusch, E.; Oehlers, S.H.; Walton, E.M.; Tram, T.T.B.; Kuang, Y.-L.; Rotter, J.I.; et al. Human genetic variation inVAC14regulatesSalmonellainvasion and typhoid fever through modulation of cholesterol. Proc. Natl. Acad. Sci. USA 2017, 114, E7746–E7755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van De Vosse, E.; Ali, S.; De Visser, A.W.; Surjadi, C.; Widjaja, S.; Vollaard, A.M.; Van Dissel, J.T. Susceptibility to typhoid fever is associated with a polymorphism in the cystic fibrosis transmembrane conductance regulator (CFTR). Hum. Genet. 2005, 118, 138–140. [Google Scholar] [CrossRef]
- Van De Vosse, E.; De Visser, A.W.; Al-Attar, S.; Vossen, R.; Ali, S.; Van Dissel, J.T. Distribution of CFTR Variations in an Indonesian Enteric Fever Cohort. Clin. Infect. Dis. 2010, 50, 1231–1237. [Google Scholar] [CrossRef]
- Dunstan, S.J.; Stephens, H.A.; Blackwell, J.M.; Duc, C.M.; Lanh, M.N.; Dudbridge, F.; Phuong, C.X.T.; Luxemburger, C.; Wain, J.; Ho, V.A.; et al. Genes of the Class II and Class III Major Histocompatibility Complex Are Associated with Typhoid Fever in Vietnam. J. Infect. Dis. 2001, 183, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Dunstan, S.J.; Hue, N.T.; Han, B.; Li, Z.; Tram, T.T.B.; Sim, K.-S.; Parry, C.M.; Chinh, N.T.; Vinh, H.; Lan, N.P.H.; et al. Variation at HLA-DRB1 is associated with resistance to enteric fever. Nat. Genet. 2014, 46, 1333–1336. [Google Scholar] [CrossRef]
- Ali, S.; Vollaard, A.; Kremer, D.; De Visser, A.W.; Martina, C.A.; Widjaja, S.; Surjadi, C.; Slagboom, E.; Van De Vosse, E.; Van Dissel, J.T.; et al. Polymorphisms in Proinflammatory Genes and Susceptibility to Typhoid Fever and Paratyphoid Fever. J. Interf. Cytokine Res. 2007, 27, 271–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunstan, S.J.; Hawn, T.R.; Nguyen, H.T.; Parry, C.P.; Ho, V.A.; Vinh, H.; Diep, T.S.; House, D.; Wain, J.; Aderem, A.; et al. Host Susceptibility and Clinical Outcomes in Toll-like Receptor 5–Deficient Patients with Typhoid Fever in Vietnam. J. Infect. Dis. 2005, 191, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sivaji, I.; Duraisamy, S.; Senthilkumar, B. Analysis of TLR polymorphisms in typhoid patients and asymptomatic typhoid carriers among the schoolchildren. Egypt. J. Med. Hum. Genet. 2016, 17, 353–357. [Google Scholar] [CrossRef] [Green Version]
- Dunstan, S.J.; Ho, V.A.; Duc, C.M.; Lanh, M.N.; Phuong, C.X.T.; Luxemburger, C.; Wain, J.; Dudbridge, F.; Peacock, C.S.; House, D.; et al. Typhoid fever and genetic polymorphisms at the natural resistance-associated macrophage protein 1. J. Infect. Dis. 2001, 183, 1156–1160. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [Green Version]
- Akira, S.; Takeda, K.; Kaisho, T. Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2001, 2, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Royle, M.C.J.; Tötemeyer, S.; Alldridge, L.C.; Maskell, D.J.; Bryant, C.E. Stimulation of Toll-Like Receptor 4 by Lipopolysaccharide During Cellular Invasion by LiveSalmonella typhimuriumIs a Critical But Not Exclusive Event Leading to Macrophage Responses. J. Immunol. 2003, 170, 5445–5454. [Google Scholar] [CrossRef] [Green Version]
- Mogensen, T.H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [Green Version]
- Poltorak, A.; He, X.; Smirnova, I.; Liu, M.-Y.; Van Huffel, C.; Du, X.; Birdwell, D.; Alejos, E.; Silva, M.; Galanos, C.; et al. Defective LPS Signaling in C3H/HeJ and C57BL/10ScCr Mice: Mutations in Tlr4 Gene. Science 1998, 282, 2085–2088. [Google Scholar] [CrossRef] [Green Version]
- Rallabhandi, P.; Bell, J.; Boukhvalova, M.S.; Medvedev, A.; Lorenz, E.; Arditi, M.; Hemming, V.G.; Blanco, J.C.G.; Segal, D.M.; Vogel, S.N. Analysis of TLR4 Polymorphic Variants: New Insights into TLR4/MD-2/CD14 Stoichiometry, Structure, and Signaling. J. Immunol. 2006, 177, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Arbour, N.C.; Lorenz, E.; Schutte, B.C.; Zabner, J.; Kline, J.N.; Jones, M.; Frees, K.; Watt, J.L.; Schwartz, D.A. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat. Genet. 2000, 25, 187–191. [Google Scholar] [CrossRef]
- Talbot, S.; Toetemeyer, S.; Yamamoto, M.; Akira, S.; Hughes, K.; Gray, D.; Barr, T.; Mastroeni, P.; Maskell, D.J.; Bryant, C.E. Toll-like receptor 4 signalling through MyD88 is essential to controlSalmonella entericaserovar Typhimurium infection, but not for the initiation of bacterial clearance. Immunology 2009, 128, 472–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnare, M.; Barton, G.M.; Holt, A.C.; Takeda, K.; Akira, S.; Medzhitov, R. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2001, 2, 947–950. [Google Scholar] [CrossRef]
- Pasare, C.; Medzhitov, R. Toll Pathway-Dependent Blockade of CD4+CD25+ T Cell-Mediated Suppression by Dendritic Cells. Science 2003, 299, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Pham, O.H.; McSorley, S.J. Protective host immune responses to Salmonella infection. Future Microbiol. 2015, 10, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Cervantes-Barragán, L.; Gil-Cruz, C.; Pastelin-Palacios, R.; Lang, K.S.; Isibasi, A.; Ludewig, B.; López-Macías, C. TLR2 and TLR4 signaling shapes specific antibody responses to Salmonella Typhi antigens. Eur. J. Immunol. 2009, 39, 126–135. [Google Scholar] [CrossRef]
- Sarma, V.N.; Malaviya, A.N.; Kumar, R.; Ghai, O.P.; Bakhtary, M.M. Development of immune response during typhoid fever in man. Clin. Exp. Immunol. 1977, 28, 35–39. [Google Scholar] [PubMed]
- Bhuiyan, S.; Sayeed, A.; Khanam, F.; Leung, D.T.; Bhuiyan, T.R.; Sheikh, A.; Salma, U.; Larocque, R.C.; Harris, J.B.; Pacek, M.; et al. Cellular and Cytokine Responses to Salmonella enterica Serotype Typhi Proteins in Patients with Typhoid Fever in Bangladesh. Am. J. Trop. Med. Hyg. 2014, 90, 1024–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamid, N.; Jain, S.K. Immunological, cellular and molecular events in typhoid fever. Indian J. Biochem. Biophys. 2007, 44, 320–330. [Google Scholar]
- Yoon, H.J.; Choi, J.Y.; Kim, C.O.; Park, Y.S.; Kim, M.S.; Kim, Y.K.; Shin, S.Y.; Kim, J.M.; Song, Y.G. Lack of Toll-like Receptor 4 and 2 Polymorphisms in Korean Patients with Bacteremia. J. Korean Med. Sci. 2006, 21, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Rezazadeh, M.; Hajilooi, M.; Rafiei, A.; Haidari, M.; Nikoopour, E.; Kerammat, F.; Mamani, M.; Ranjbar, M.; Hashemi, H. TLR4 polymorphism in Iranian patients with brucellosis. J. Infect. 2006, 53, 206–210. [Google Scholar] [CrossRef]
- Gazouli, M.; Mantzaris, G.; Kotsinas, A.; Zacharatos, P.; Papalambros, E.; Archimandritis, A.; Ikonomopoulos, J.; Gorgoulis, V.G. Association between polymorphisms in the Toll-like receptor 4, CD14, andCARD15/NOD2and inflammatory bowel disease in the Greek population. World J. Gastroenterol. 2005, 11, 681–685. [Google Scholar] [CrossRef]
- Okayama, N.; Fujimura, K.; Suehiro, Y.; Hamanaka, Y.; Fujiwara, M.; Matsubara, T.; Maekawa, T.; Hazama, S.; Oka, M.; Nohara, H.; et al. Simple genotype analysis of the Asp299Gly polymorphism of the Toll-like receptor-4 gene that is associated with lipopolysaccharide hyporesponsiveness. J. Clin. Lab. Anal. 2002, 16, 56–58. [Google Scholar] [CrossRef]
- Hue, N.T.; Lanh, M.N.; Phuong, L.T.; Vinh, H.; Chinh, N.T.; Hien, T.T.; Hieu, N.T.; Farrar, J.J.; Dunstan, S.J. Toll-Like Receptor 4 (TLR4) and Typhoid Fever in Vietnam. PLoS ONE 2009, 4, e4800. [Google Scholar] [CrossRef] [Green Version]
- Mastroeni, P.; Sheppard, M. Salmonella infections in the mouse model: Host resistance factors and in vivo dynamics of bacterial spread and distribution in the tissues. Microbes Infect. 2004, 6, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, E.; Mira, J.P.; Frees, K.L.; Schwartz, D.A. Relevance of Mutations in the TLR4 Receptor in Patients with Gram-Negative Septic Shock. Arch. Intern. Med. 2002, 162, 1028–1032. [Google Scholar] [CrossRef] [PubMed]
- Sultzer, B.M. Genetic Control of Leucocyte Responses to Endotoxin. Nature 1968, 219, 1253–1254. [Google Scholar] [CrossRef]
- Georgel, P.; Macquin, C.; Bahram, S. The Heterogeneous Allelic Repertoire of Human Toll-Like Receptor (TLR) Genes. PLoS ONE 2009, 4, e7803. [Google Scholar] [CrossRef] [Green Version]
- Papadopoulos, A.; Ferwerda, B.; Antoniadou, A.; Sakka, V.; Galani, L.; Kavatha, D.; Panagopoulos, P.; Poulakou, G.; Kanellakopoulou, K.; Van Der Meer, J.W.M.; et al. Association of Toll-Like Receptor 4 Asp299Gly and Thr399Ile Polymorphisms with Increased Infection Risk in Patients with Advanced HIV-1 Infection. Clin. Infect. Dis. 2010, 51, 242–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gianchecchi, E.; Torelli, A.; Piccini, G.; Piccirella, S.; Montomoli, E. N. meningitidis and TLR Polymorphisms: A Fascinating Immunomodulatory Network. Vaccines 2016, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Mastroeni, P.; Vazquez-Torres, A.; Fang, F.C.; Xu, Y.; Khan, S.; Hormaeche, C.E.; Dougan, G. Antimicrobial Actions of the Nadph Phagocyte Oxidase and Inducible Nitric Oxide Synthase in Experimental Salmonellosis. II. Effects on Microbial Proliferation and Host Survival in Vivo. J. Exp. Med. 2000, 192, 237–248. [Google Scholar] [CrossRef]
- Erridge, C.; Stewart, J.; Poxton, I.R. Monocytes Heterozygous for the Asp299Gly and Thr399Ile Mutations in the Toll-like Receptor 4 Gene Show No Deficit in Lipopolysaccharide Signalling. J. Exp. Med. 2003, 197, 1787–1791. [Google Scholar] [CrossRef] [PubMed]
- Hawn, T.R.; Verbon, A.; Janer, M.; Zhao, L.P.; Beutler, B.; Aderem, A. Toll-like receptor 4 polymorphisms are associated with resistance to Legionnaires’ disease. Proc. Natl. Acad. Sci. USA 2005, 102, 2487–2489. [Google Scholar] [CrossRef] [Green Version]
- Smirnova, I.; Mann, N.; Dols, A.; Derkx, H.H.; Hibberd, M.L.; Levin, M.; Beutler, B. Assay of locus-specific genetic load implicates rare Toll-like receptor 4 mutations in meningococcal susceptibility. Proc. Natl. Acad. Sci. USA 2003, 100, 6075–6080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schröder, N.W.J.; Schumann, R.R. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect. Dis. 2005, 5, 156–164. [Google Scholar] [CrossRef]
- Mockenhaupt, F.P.; Cramer, J.P.; Hamann, L.; Stegemann, M.S.; Eckert, J.; Oh, N.-R.; Otchwemah, R.N.; Dietz, E.; Ehrhardt, S.; Schröder, N.W.J.; et al. Toll-like receptor (TLR) polymorphisms in African children: Common TLR-4 variants predispose to severe malaria. Proc. Natl. Acad. Sci. USA 2006, 103, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Ferwerda, B.; McCall, M.; Alonso, S.; Giamarellos-Bourboulis, E.J.; Mouktaroudi, M.; Izagirre, N.; Syafruddin, D.; Kibiki, G.; Cristea, T.; Hijmans, A.; et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc. Natl. Acad. Sci. USA 2007, 104, 16645–16650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kröner, A.; Vogel, F.; Kolb-Mäurer, A.; Kruse, N.; Toyka, K.; Hemmer, B.; Rieckmann, P.; Maurer, M. Impact of the Asp299Gly polymorphism in the toll-like receptor 4 (tlr-4) gene on disease course of multiple sclerosis. J. Neuroimmunol. 2005, 165, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Misch, E.A.; Hawn, T.R. Toll-like receptor polymorphisms and susceptibility to human disease. Clin. Sci. 2008, 114, 347–360. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, F.; Smith, K.D.; Ozinsky, A.; Hawn, T.R.; Yi, E.C.; Goodlett, D.R.; Eng, J.K.; Akira, S.; Underhill, D.M.; Aderem, A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001, 410, 1099–1103. [Google Scholar] [CrossRef]
- McSorley, S.J.; Ehst, B.D.; Yu, Y.; Gewirtz, A.T. Bacterial Flagellin Is an Effective Adjuvant for CD4+T Cells In Vivo. J. Immunol. 2002, 169, 3914–3919. [Google Scholar] [CrossRef] [Green Version]
- Chaichana, P.; Chantratita, N.; Brod, F.; Koosakulnirand, S.; Jenjaroen, K.; Chumseng, S.; Sumonwiriya, M.; Burtnick, M.N.; Brett, P.J.; Teparrukkul, P.; et al. A nonsense mutation in TLR5 is associated with survival and reduced IL-10 and TNF-α levels in human melioidosis. PLoS Negl. Trop. Dis. 2017, 11, e0005587. [Google Scholar] [CrossRef] [Green Version]
- Fournier, B.; Williams, I.R.; Gewirtz, A.T.; Neish, A.S. Toll-Like Receptor 5-Dependent Regulation of Inflammation in Systemic Salmonella enterica Serovar Typhimurium Infection. Infect. Immun. 2009, 77, 4121–4129. [Google Scholar] [CrossRef] [Green Version]
- Hawn, T.R.; Verbon, A.; Lettinga, K.D.; Zhao, L.P.; Li, S.S.; Laws, R.J.; Skerrett, S.J.; Beutler, B.; Schroeder, L.; Nachman, A.; et al. A Common Dominant TLR5 Stop Codon Polymorphism Abolishes Flagellin Signaling and Is Associated with Susceptibility to Legionnaires’ Disease. J. Exp. Med. 2003, 198, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Hawn, T.R.; Wu, H.; Grossman, J.M.; Hahn, B.H.; Tsao, B.P.; Aderem, A. A stop codon polymorphism of Toll-like receptor 5 is associated with resistance to systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 2005, 102, 10593–10597. [Google Scholar] [CrossRef] [Green Version]
- Senthilkumar, B.; Sivakumar, P.; Madhanraj, R.; Senbagam, D.; Illakia, S. A comparative analysis of TLR5 polymorphism and clinical parameters in typhoid patients and asymptomatic typhoid carriers. J. Public Health 2014, 22, 131–137. [Google Scholar] [CrossRef]
- Canonne-Hergaux, F.; Samantha, G.; Gregory, G.; Philippe, G. The Nramp1 protein and its role in resistance to infection and macrophage function. Proc. Assoc. Am. Physicians 2003, 111, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Y.; Zhou, F.; Zhang, Y.; Lu, H.; Jin, Q.; Gao, L. SLC11A1 (NRAMP1) Polymorphisms and Tuberculosis Susceptibility: Updated Systematic Review and Meta-Analysis. PLoS ONE 2011, 6, e15831. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.S.; Kim, S.J.; Kim, J.W.; Koh, E.M. NRAMP1gene polymorphisms in patients with rheumatoid arthritis in Koreans. J. Korean Med. Sci. 2000, 15, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Cunrath, O.; Bumann, D. Host resistance factor SLC11A1 restricts Salmonella growth through magnesium deprivation. Science 2019, 366, 995–999. [Google Scholar] [CrossRef] [PubMed]
- Ateş, Ö.; Dalyan, L.; Müsellim, B.; Hatemi, G.; Türker, H.; Öngen, G.; Hamuryudan, V.; Topal-Sarıkaya, A. NRAMP1 (SLC11A1) gene polymorphisms that correlate with autoimmune versus infectious disease susceptibility in tuberculosis and rheumatoid arthritis. Int. J. Immunogenet. 2009, 36, 15–19. [Google Scholar] [CrossRef]
- Gazouli, M.; Atsaves, V.; Mantzaris, G.; Economou, M.; Nasioulas, G.; Evangelou, K.; Archimandritis, A.J.; Anagnou, N.P. Role of functional polymorphisms of NRAMP1 gene for the development of Crohn’s disease. Inflamm. Bowel Dis. 2008, 14, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Medapati, R.V.; Suvvari, S.; Sudhakar, G.; Gangisetti, P. NRAMP1 and VDR gene polymorphisms in susceptibility to pulmonary tuberculosis among Andhra Pradesh population in India: A case–control study. BMC Pulm. Med. 2017, 17, 89. [Google Scholar] [CrossRef] [PubMed]
- Brochado, M.J.F.; Gatti, M.F.C.; Zago, M.A.; Roselino, A.M. Association of the solute carrier family 11 member 1 gene polymorphisms with susceptibility to leprosy in a Brazilian sample. Mem. Inst. Oswaldo Cruz 2016, 111, 101–105. [Google Scholar] [CrossRef]
- Schulze, U.; Vollenbröker, B.; Braun, D.A.; Van Le, T.; Granado, D.; Kremerskothen, J.; Fränzel, B.; Klosowski, R.; Barth, J.; Fufezan, C.; et al. The Vac14-interaction Network Is Linked to Regulators of the Endolysosomal and Autophagic Pathway. Mol. Cell. Proteom. 2014, 13, 1397–1411. [Google Scholar] [CrossRef] [Green Version]
- Jin, N.; Chow, C.Y.; Liu, L.; Zolov, S.N.; Bronson, R.; Davisson, M.; Petersen, J.L.; Zhang, Y.; Park, S.; Duex, J.E.; et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P2 in yeast and mouse. EMBO J. 2008, 27, 3221–3234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilchrist, J.J.; Mentzer, A.J.; Rautanen, A.; Pirinen, M.; Mwarumba, S.; Njuguna, P.; Mturi, N.; Wellcome Trust Case-Control Consortium 2; The Kenyan Bacteraemia Study Group; Williams, T.N.; et al. Genetic variation in VAC14 is associated with bacteremia secondary to diverse pathogens in African children. Proc. Natl. Acad. Sci. USA 2018, 115, E3601–E3603. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, M.I.; Ko, D.C. Reply to Gilchrist et al.: Possible roles for VAC14 in multiple infectious diseases. Proc. Natl. Acad. Sci. USA 2018, 115, E3604–E3605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Duan, Q.; Li, Y.; Yang, Y.; Hardwidge, P.R.; Zhu, G. Membrane cholesterol plays an important role in enteropathogen adhesion and the activation of innate immunity via flagellin–TLR5 signaling. Arch. Microbiol. 2015, 197, 797–803. [Google Scholar] [CrossRef]
- Jin, J.S.; Kwon, S.-O.; Moon, D.C.; Gurung, M.; Lee, J.H.; Kim, S.I.; Lee, J.C. Acinetobacter baumannii Secretes Cytotoxic Outer Membrane Protein A via Outer Membrane Vesicles. PLoS ONE 2011, 6, e17027. [Google Scholar] [CrossRef]
- Gradstedt, H.; Iovino, F.; Bijlsma, J.J.E. Streptococcus pneumoniae Invades Endothelial Host Cells via Multiple Pathways and Is Killed in a Lysosome Dependent Manner. PLoS ONE 2013, 8, e65626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blohmke, C.J.; Darton, T.C.; Jones, C.; Suarez, N.M.; Waddington, C.S.; Angus, B.; Zhou, L.; Hill, J.; Clare, S.; Kane, L.; et al. Interferon-driven alterations of the host’s amino acid metabolism in the pathogenesis of typhoid fever. J. Exp. Med. 2016, 213, 1061–1077. [Google Scholar] [CrossRef] [PubMed]
- Owen, K.A.; Anderson, C.J.; Casanova, J.E. Salmonella Suppresses the TRIF-Dependent Type I Interferon Response in Macrophages. mBio 2016, 7, e02051–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotolongo, J.; España, C.; Echeverry, A.; Siefker, D.; Altman, N.; Zaias, J.; Santaolalla, R.; Ruiz, J.; Schesser, K.; Adkins, B.; et al. Host innate recognition of an intestinal bacterial pathogen induces TRIF-dependent protective immunity. J. Exp. Med. 2011, 208, 2705–2716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, F.-C.; Young, H.A. Interferons: Success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 2014, 25, 369–376. [Google Scholar] [CrossRef] [Green Version]
- Sheikh, A.; Khanam, F.; Sayeed, A.; Rahman, T.; Pacek, M.; Hu, Y.; Rollins, A.; Bhuiyan, S.; Rollins, S.; Kalsy, A.; et al. Interferon-γ and Proliferation Responses to Salmonella enterica Serotype Typhi Proteins in Patients with S. Typhi Bacteremia in Dhaka, Bangladesh. PLoS Neglected Trop. Dis. 2011, 5, e1193. [Google Scholar] [CrossRef]
- Butler, T.; Ho, M.; Acharya, G.; Tiwari, M.; Gallati, H. Interleukin-6, gamma interferon, and tumor necrosis factor receptors in typhoid fever related to outcome of antimicrobial therapy. Antimicrob. Agents Chemother. 1993, 37, 2418–2421. [Google Scholar] [CrossRef] [Green Version]
- Jouanguy, E.; Dupuis, S.; Pallier, A.; Döffinger, R.; Fondanèche, M.-C.; Fieschi, C.; Lamhamedi-Cherradi, S.; Altare, F.; Emile, J.-F.; Lutz, P.; et al. In a novel form of IFN-γ receptor 1 deficiency, cell surface receptors fail to bind IFN-γ. J. Clin. Investig. 2000, 105, 1429–1436. [Google Scholar] [CrossRef] [Green Version]
- Nairz, M.; Fritsche, G.; Brunner, P.; Talasz, H.; Hantke, K.; Weiss, G. Interferon-γ limits the availability of iron for intramacrophage Salmonella Typhimurium. Eur. J. Immunol. 2008, 38, 1923–1936. [Google Scholar] [CrossRef]
- Nairz, M.; Haschka, D.; Demetz, E.; Weiss, G. Iron at the interface of immunity and infection. Front. Pharmacol. 2014, 5, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govoni, G.; Gauthier, S.; Billia, F.; Iscove, N.N.; Gros, P. Cell-specific and inducible Nramp1 gene expression in mouse macrophages in vitro and in vivo. J. Leukoc. Biol. 1997, 62, 277–286. [Google Scholar] [CrossRef]
- Fritsche, G.; Nairz, M.; Libby, S.J.; Fang, F.C.; Weiss, G. Slc11a1 (Nramp1) impairs growth of Salmonella enterica serovar Typhimuriumin macrophages via stimulation of lipocalin-2 expression. J. Leukoc. Biol. 2012, 92, 353–359. [Google Scholar] [CrossRef] [Green Version]
- MacMicking, J.D. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 2012, 12, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Kagaya, K.; Watanabe, K.; Fukazawa, Y. Capacity of recombinant gamma interferon to activate macrophages for Salmonella-killing activity. Infect. Immun. 1989, 57, 609–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birmingham, C.L.; Smith, A.C.; Bakowski, M.A.; Yoshimori, T.; Brumell, J.H. Autophagy Controls Salmonella Infection in Response to Damage to the Salmonella-containing Vacuole. J. Biol. Chem. 2006, 281, 11374–11383. [Google Scholar] [CrossRef] [Green Version]
- Gilchrist, J.J.; MacLennan, C.A.; Hill, A.V.S. Genetic susceptibility to invasive Salmonella disease. Nat. Rev. Immunol. 2015, 15, 452–463. [Google Scholar] [CrossRef]
- Nakamura, Y.; Koyama, K.; Matsushima, M. VNTR (variable number of tandem repeat) sequences as transcriptional, translational, or functional regulators. J. Hum. Genet. 1998, 43, 149–152. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, B.; Martinson, M.; Webb, G.; Young, I. Molecular organization of the cytokine gene cluster, involving the human IL-3, IL-4, IL-5, and GM-CSF genes, on human chromosome 5. Blood 1989, 73, 1142–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunal, O.; Yigit, S.; Yalcın, A.D.; Celik, B.; Barut, S.; Demir, O.; Ates, O.; Duygu, F.; Kaya, S.; Rüstemoğlu, A.; et al. The IL4-VNTR P1 Allele, IL4-VNTR P2P2 Genotype, and IL4-VNTR_IL6-174CG P2P1-GG Genotype Are Associated with an Increased Risk of Brucellosis. Jpn. J. Infect. Dis. 2017, 70, 61–64. [Google Scholar] [CrossRef]
- Yigit, S.; Inanir, A.; Tekcan, A.; Tural, E.; Ozturk, G.T.; Kismali, G.; Karakuş, N.; Tekcan, A. Significant association of interleukin-4 gene intron 3 VNTR polymorphism with susceptibility to knee osteoarthritis. Gene 2014, 537, 6–9. [Google Scholar] [CrossRef]
- Karakus, N.; Yigit, S.; Kurt, G.S.; Cevik, B.; Demir, O.; Ateş, Ö. Association of interleukin (IL)-4 gene intron 3 VNTR polymorphism with multiple sclerosis in Turkish population. Hum. Immunol. 2013, 74, 1157–1160. [Google Scholar] [CrossRef]
- Sood, S.; Rishi, P.; Vohra, H.; Sharma, S.; Ganguly, N.K. Cellular immune response induced by Salmonella enterica serotype Typhi iron-regulated outer-membrane proteins at peripheral and mucosal levels. J. Med. Microbiol. 2005, 54, 815–821. [Google Scholar] [CrossRef] [Green Version]
- Ley, K. M1 Means Kill; M2 Means Heal. J. Immunol. 2017, 199, 2191–2193. [Google Scholar] [CrossRef]
- Italiani, P.; Boraschi, D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 2014, 5, 514. [Google Scholar] [CrossRef] [Green Version]
- Luzina, I.G.; Keegan, A.D.; Heller, N.M.; Rook, G.A.W.; Shea-Donohue, T.; Atamas, S.P. Regulation of inflammation by interleukin-4: A review of “alternatives”. J. Leukoc. Biol. 2012, 92, 753–764. [Google Scholar] [CrossRef] [Green Version]
- Chopra, R.; Ali, S.; Srivastava, A.K.; Aggarwal, S.; Kumar, B.; Manvati, S.; Kalaiarasan, P.; Jena, M.; Garg, V.K.; Bhattacharya, S.N.; et al. Mapping of PARK2 and PACRG Overlapping Regulatory Region Reveals LD Structure and Functional Variants in Association with Leprosy in Unrelated Indian Population Groups. PLoS Genet. 2013, 9, e1003578. [Google Scholar] [CrossRef] [Green Version]
- Shimura, H.; Hattori, N.; Kubo, S.-I.; Mizuno, Y.; Asakawa, S.; Minoshima, S.; Shimizu, N.; Iwai, K.; Chiba, T.; Tanaka, K.; et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 2000, 25, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.C.; Hueffer, K.; Lam, T.T.; Galán, J.E. Diversification of a Salmonella Virulence Protein Function by Ubiquitin-Dependent Differential Localization. Cell 2009, 137, 283–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Higashide, W.; Dai, S.; Sherman, D.M.; Zhou, D. Recognition and Ubiquitination of Salmonella Type III Effector SopA by a Ubiquitin E3 Ligase, HsRMA1. J. Biol. Chem. 2005, 280, 38682–38688. [Google Scholar] [CrossRef] [Green Version]
- Kubori, T.; Galán, J.E. Temporal Regulation of Salmonella Virulence Effector Function by Proteasome-Dependent Protein Degradation. Cell 2003, 115, 333–342. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Galán, J.E. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 1999, 401, 293–297. [Google Scholar] [CrossRef]
- Zohaib, A.; Duan, X.; Zhu, B.; Ye, J.; Wan, S.; Chen, H.; Liu, X.; Cao, S. The Role of Ubiquitination in Regulation of Innate Immune Signaling. Curr. Issues Mol. Biol. 2015, 18, 1–10. [Google Scholar] [PubMed]
- De Léséleuc, L.; Orlova, M.; Cobat, A.; Girard, M.; Huong, N.T.; Ba, N.N.; Van Thuc, N.; Truman, R.; Spencer, J.S.; Adams, L.; et al. PARK2 Mediates Interleukin 6 and Monocyte Chemoattractant Protein 1 Production by Human Macrophages. PLoS Negl. Trop. Dis. 2013, 7, e2015. [Google Scholar] [CrossRef] [PubMed]
- Khaminets, A.; Behl, C.; Dikic, I. Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 2016, 26, 6–16. [Google Scholar] [CrossRef]
- Cohen, P. Immune diseases caused by mutations in kinases and components of the ubiquitin system. Nat. Immunol. 2014, 15, 521–529. [Google Scholar] [CrossRef] [Green Version]
- McEwan, D.G. Host–pathogen interactions and subversion of autophagy. Essays Biochem. 2017, 61, 687–697. [Google Scholar] [CrossRef] [Green Version]
- Manzanillo, P.S.; Ayres, J.S.; Watson, R.O.; Collins, A.C.; Souza, G.; Rae, C.S.; Schneider, D.S.; Nakamura, K.; Shiloh, M.U.; Cox, J.S. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 2013, 501, 512–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heath, R.J.; Goel, G.; Baxt, L.A.; Rush, J.S.; Mohanan, V.; Paulus, G.L.; Jani, V.; Lassen, K.G.; Xavier, R.J. RNF166 Determines Recruitment of Adaptor Proteins during Antibacterial Autophagy. Cell Rep. 2016, 17, 2183–2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.-L.; Wu, Y.-W.; Kuo, C.-F.; Lu, S.-L.; Liu, F.-T.; Anderson, R.; Lin, C.-F.; Liu, Y.-L.; Wang, W.-Y.; Chen, Y.-D.; et al. Galectin-3 Inhibits Galectin-8/Parkin-Mediated Ubiquitination of Group A Streptococcus. mBio 2017, 8, e00899-17. [Google Scholar] [CrossRef] [Green Version]
- Mira, M.T.; Alcaïs, A.; Van Thuc, N.; Moraes, M.O.; Di Flumeri, C.; Thai, V.H.; Phuong, M.C.; Huong, N.T.; Ba, N.N.; Khoa, P.X.; et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature 2004, 427, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Alter, A.; Fava, V.M.; Huong, N.T.; Singh, M.; Orlova, M.; Van Thuc, N.; Katoch, K.; Thai, V.H.; Ba, N.N.; Abel, L.; et al. Linkage disequilibrium pattern and age-at-diagnosis are critical for replicating genetic associations across ethnic groups in leprosy. Hum. Genet. 2013, 132, 107–116. [Google Scholar] [CrossRef]
- Hanrahan, J.W.; Wioland, M.A. Revisiting cystic fibrosis transmembrane conductance regulator structure and function. Proc. Am. Thorac. Soc. 2004, 1, 17–21. [Google Scholar] [CrossRef]
- Bobadilla, J.L.; Macek, M.; Fine, J.P.; Farrell, P.M. Cystic fibrosis: A worldwide analysis of CFTR mutations-correlation with incidence data and application to screening. Hum. Mutat. 2002, 19, 575–606. [Google Scholar] [CrossRef]
- Estivill, X.; Bancells, C.; Ramos, C. Geographic distribution and regional origin of 272 cystic fibrosis mutations in European populations. The Biomed CF Mutation Analysis Consortium. Hum. Mutat. 1997, 10, 135–154. [Google Scholar] [CrossRef]
- Niel, F.; Martin, J.; Moal, F.D.-L.; Costes, B.; Boissier, B.; Delattre, V.; Goossens, M.; Girodon, E. Rapid detection of CFTR gene rearrangements impacts on genetic counselling in cystic fibrosis. J. Med. Genet. 2004, 41, e118. [Google Scholar] [CrossRef]
- Pier, G.B.; Grout, M.; Zaidi, T.M.; Meluleni, G.; Mueschenborn, S.S.; Banting, G.; Ratcliff, R.; Evans, M.J.; Colledge, W.H. Salmonella Typhi uses CFTR to enter intestinal epithelial cells. Nature 1998, 393, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Lyczak, J.B.; Pier, G.B. Salmonella enterica Serovar Typhi Modulates Cell Surface Expression of Its Receptor, the Cystic Fibrosis Transmembrane Conductance Regulator, on the Intestinal Epithelium. Infect. Immun. 2002, 70, 6416–6423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoß, M.; Ilmonen, P.; Musolf, K.; Penn, D.J. Major histocompatibility complex heterozygosity enhances reproductive success. Mol. Ecol. 2011, 20, 1546–1557. [Google Scholar] [CrossRef]
- Woelfing, B.; Traulsen, A.; Milinski, M.; Boehm, T. Does intra-individual major histocompatibility complex diversity keep a golden mean? Philos. Trans. R. Soc. B: Biol. Sci. 2009, 364, 117–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.; Adams, S.; Marincola, F.M.; Stroncek, D.F. Human Leukocyte and Granulocyte Antigens and Antibodies: The HLA and HNA Systems A2—Hillyer, Christopher D. In Blood Banking and Transfusion Medicine, 2nd ed.; Silberstein, L.E., Ness, P.M., Anderson, K.C., Roback, J.D., Eds.; Churchill Livingstone: Philadelphia, PA, USA, 2007; Chapter 10; pp. 129–156. [Google Scholar]
- Blackwell, J.M.; Jamieson, S.E.; Burgner, D. HLA and Infectious Diseases. Clin. Microbiol. Rev. 2009, 22, 370–385. [Google Scholar] [CrossRef] [Green Version]
- Pollack, M.S.; Rich, R.R. The HLA Complex and the Pathogenesis of Infectious Diseases. J. Infect. Dis. 1985, 151, 1–8. [Google Scholar] [CrossRef]
- Tian, C.; Hromatka, B.S.; Kiefer, A.K.; Eriksson, N.; Noble, S.M.; Tung, J.Y.; Hinds, D. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Crosslin, D.R.; Carrell, D.S.; Burt, A.; Kim, D.S.; Underwood, J.G.; Hanna, D.S.; Comstock, B.A.; Baldwin, E.; De Andrade, M.; Kullo, I.J.; et al. Genetic variation in the HLA region is associated with susceptibility to herpes zoster. Genes Immun. 2015, 16, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dharmana, E.; Joosten, I.; Tijssen, H.J.; Gasem, M.H.; Indarwidayati, R.; Keuter, M.; Dolmans, W.M.V.; Van Der Meer, J.W.M. HLA-DRB1*12 is associated with protection against complicated typhoid fever, independent of tumour necrosis factor alpha. Eur. J. Immunogenet. 2002, 29, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Elahi, M.M.; Asotra, K.; Matata, B.M.; Mastana, S.S. Tumor necrosis factor alpha −308 gene locus promoter polymorphism: An analysis of association with health and disease. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2009, 1792, 163–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene a | Location of Gene | Influence of Variants | Reference |
|---|---|---|---|
| Gene Variants that Affect the Enteric Fever Outcomes | |||
| TLR4 | Chromosome 9 | Missense mutation in TLR4 (threonine → isoleucine substitution at position 399 of the amino acid sequence) or (aspartate → glycine substitution at position 299 of the amino acid sequence) is evidenced to be associated with an increasing risk for Salmonella infection and severity of enteric fever. | [13,14] |
| IL-4 | Chromosome 5 | Variable number of tandem repeat polymorphisms of 3R2R at IL-4 could be a genetic predisposition factor for S. Typhi or S. Paratyphi infection. | [15] |
| VAC14 | Chromosome 16 | The polymorphism rs8060947 in VAC14 gene renders it susceptible to infection. | [16] |
| PARK2/PACRG | Chromosome 6 | Mutation in PARK2 results in a single-nucleotide polymorphism of PARK2_e01(−2599) which shows the weak association and susceptibility to typhoid fever and paratyphoid fever. | [12] |
| CFTR | Chromosome 7 | Polymorphic dinucleotide repeats in the intron or exon of the CFTR gene and also in the single nucleotide variant, whereas, polymorphisms poly-T at CFTR gene are found to be associated with protection against enteric fever. | [17,18] |
| HLAGene Complex Class II and Class III | Chromosome 6 | HLA-DRB1*04:05 and TNF*1 (−308) allele is associated with resistance to enteric fever whereas HLA-DRB1*0301/6/8 and HLA-DQB1*0201-3 allele are associated with susceptibility to enteric fever. TNFA*2 (−308) is associated with the outcome of Salmonella typhoidal species infection. | [19,20,21] |
| Gene Variants that Do Not Affect the Enteric Fever Outcomes | |||
| TLR5 | Chromosome 1 | TLR5 variants do not have a significant effect on the susceptibility or severity of enteric fever. | [22,23] |
| NRAMP1 | Chromosome 2 | NRAMP1 polymorphisms are not associated with acquiring enteric fever. | [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, P.Y.; Tan, J.E.; Hee, E.W.; Yong, D.W.X.; Heng, Y.S.; Low, W.X.; Wu, X.H.; Cletus, C.; Kumar Chellappan, D.; Aung, K.; et al. Human Genetic Variation Influences Enteric Fever Progression. Cells 2021, 10, 345. https://doi.org/10.3390/cells10020345
Ma PY, Tan JE, Hee EW, Yong DWX, Heng YS, Low WX, Wu XH, Cletus C, Kumar Chellappan D, Aung K, et al. Human Genetic Variation Influences Enteric Fever Progression. Cells. 2021; 10(2):345. https://doi.org/10.3390/cells10020345
Chicago/Turabian StyleMa, Pei Yee, Jing En Tan, Edd Wyn Hee, Dylan Wang Xi Yong, Yi Shuan Heng, Wei Xiang Low, Xun Hui Wu, Christy Cletus, Dinesh Kumar Chellappan, Kyan Aung, and et al. 2021. "Human Genetic Variation Influences Enteric Fever Progression" Cells 10, no. 2: 345. https://doi.org/10.3390/cells10020345
APA StyleMa, P. Y., Tan, J. E., Hee, E. W., Yong, D. W. X., Heng, Y. S., Low, W. X., Wu, X. H., Cletus, C., Kumar Chellappan, D., Aung, K., Yong, C. Y., & Liew, Y. K. (2021). Human Genetic Variation Influences Enteric Fever Progression. Cells, 10(2), 345. https://doi.org/10.3390/cells10020345