Metal Ions Induce Liquid Condensate Formation by the F Domain of Aedes aegypti Ecdysteroid Receptor. New Perspectives of Nuclear Receptor Studies
Abstract
:1. Characteristics of NRs
2. Transcription Factors in the Cellular Response to Mosquito-Borne Infections
2.1. Human Cellular Response to Viral Infection Involves Changes in Gene Expression
2.2. Transcription Factors Involved in the Mosquito’s Response to Pathogens
3. Liquid-Liquid Phase Separation in Gene Expression
Liquid-Liquid Phase Separation in Transcriptional Regulation by NRs
4. F Domains of Nuclear Receptors
4.1. F Domain of the Ecdysteroid Receptor (EcR) from A. aegypti
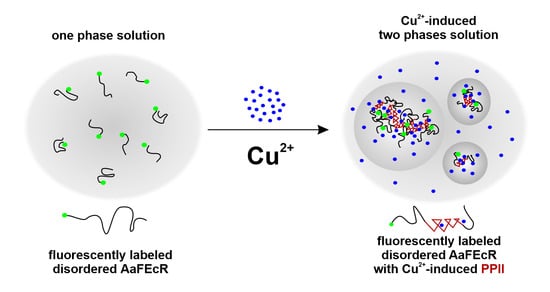
4.2. AaFEcR Provokes LLPS
5. Summary and Perspectives on Future Research
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aranda, A.; Pascual, A. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001, 81, 1269–1304. [Google Scholar] [CrossRef]
- Altucci, L.; Gronemeyer, H. Nuclear receptors in cell life and death. Trends Endocrinol. Metab. 2001, 12, 460–468. [Google Scholar] [CrossRef]
- Aagaard, M.M.; Siersbæk, R.; Mandrup, S. Molecular basis for gene-specific transactivation by nuclear receptors. Biochim. Biophys. Acta Mol. Basis Dis. 2011, 1812, 824–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawlak, M.; Lefebvre, P.; Staels, B. General molecular biology and architecture of nuclear receptors. Curr. Top. Med. Chem. 2012, 12, 486–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mcewan, I.J.; Kumar, R. Nuclear Receptors: From Structure to the Clinic; Springer: Berlin, Germany, 2015; ISBN 978-3-319-18728-0. [Google Scholar]
- Schulman, I.G. Nuclear receptors as drug targets for metabolic disease. Adv. Drug Deliv. Rev. 2010, 62, 1307–1315. [Google Scholar] [CrossRef] [Green Version]
- Weikum, E.R.; Liu, X.; Ortlund, E.A. The nuclear receptor superfamily: A structural perspective. Protein Sci. 2018, 27, 1876–1892. [Google Scholar] [CrossRef] [PubMed]
- Rastinejad, F.; Huang, P.; Chandra, V.; Khorasanizadeh, S. Understanding nuclear receptor form and function using structural biology. J. Mol. Endocrinol. 2013, 51, T1–T21. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.; Chandra, V.; Rastinejad, F. Structural Overview of the Nuclear Receptor Superfamily: Insights into Physiology and Therapeutics. Annu. Rev. Physiol. 2010, 72, 247–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rowińska-Żyrek, M.; Więch, A.; Wątły, J.; Wieczorek, R.; Witkowska, D.; Ożyhar, A.; Orłowski, M. Copper(II)-binding induces a unique polyproline type II helical structure within the ion-binding segment in the intrinsically disordered F-domain of ecdysteroid receptor from Aedes aegypti. Inorg. Chem. 2019, 58, 11782–11792. [Google Scholar] [CrossRef] [PubMed]
- Więch, A.; Rowińska-Żyrek, M.; Wątły, J.; Czarnota, A.; Hołubowicz, R.; Szewczuk, Z.; Ożyhar, A.; Orłowski, M. The intrinsically disordered C-terminal F domain of the ecdysteroid receptor from Aedes aegypti exhibits metal ion-binding ability. J. Steroid Biochem. Mol. Biol. 2019, 186, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Nocula-Ługowska, M.; Rymarczyk, G.; Lisowski, M.; Ożyhar, A. Isoform-specific variation in the intrinsic disorder of the ecdysteroid receptor N-terminal domain. Proteins Struct. Funct. Bioinform. 2009, 76, 291–308. [Google Scholar] [CrossRef]
- Pieprzyk, J.; Zbela, A.; Jakób, M.; Ożyhar, A.; Orłowski, M. Homodimerization propensity of the intrinsically disordered N-terminal domain of Ultraspiracle from Aedes aegypti. Biochim. Biophys. Acta Proteins Proteom. 2014, 1844, 1153–1166. [Google Scholar] [CrossRef]
- Wärnmark, A.; Wikström, A.; Wright, A.P.H.; Gustafsson, J.Å.; Härd, T. The N-terminal Regions of Estrogen Receptor α and β Are Unstructured in Vitro and Show Different TBP Binding Properties. J. Biol. Chem. 2001, 276, 45939–45944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, J.; Kelly, S.M.; Watt, K.; Price, N.C.; McEwan, I.J. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J. Biol. Chem. 2002, 277, 20079–20086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziedzic-Letka, A.; Rymarczyk, G.; Kapłon, T.M.; Górecki, A.; Szamborska-Gbur, A.; Wojtas, M.; Dobryszycki, P.; Ożyhar, A. Intrinsic disorder of Drosophila melanogaster hormone receptor 38 N-terminal domain. Proteins Struct. Funct. Bioinform. 2011, 79, 376–392. [Google Scholar] [CrossRef] [PubMed]
- Belorusova, A.Y.; Osz, J.; Petoukhov, M.V.; Peluso-Iltis, C.; Kieffer, B.; Svergun, D.I.; Rochel, N. Solution Behavior of the Intrinsically Disordered N-Terminal Domain of Retinoid X Receptor α in the Context of the Full-Length Protein. Biochemistry 2016, 55, 1741–1748. [Google Scholar] [CrossRef] [Green Version]
- Wycisk, K.; Tarczewska, A.; Kaus-Drobek, M.; Dadlez, M.; Hołubowicz, R.; Pietras, Z.; Dziembowski, A.; Taube, M.; Kozak, M.; Orłowski, M.; et al. Intrinsically disordered N-terminal domain of the Helicoverpa armigera Ultraspiracle stabilizes the dimeric form via a scorpion-like structure. J. Steroid Biochem. Mol. Biol. 2018, 183, 167–183. [Google Scholar] [CrossRef]
- Sołtys, K.; Ożyhar, A. Ordered structure-forming properties of the intrinsically disordered AB region of hRXRγ and its ability to promote liquid-liquid phase separation. J. Steroid Biochem. Mol. Biol. 2020, 198, 105571. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [Green Version]
- Simons, S.S.; Edwards, D.P.; Kumar, R. Minireview: Dynamic Structures of Nuclear Hormone Receptors: New Promises and Challenges. Mol. Endocrinol. 2014, 28, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Perumal, N.B.; Oldfield, C.J.; Su, E.W.; Uversky, V.N.; Dunker, A.K. Intrinsic Disorder in Transcription Factors. Biochemistry 2006, 45, 6873–6888. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Uversky, V.N. Drugs for ‘protein clouds’: Targeting intrinsically disordered transcription factors. Curr. Opin. Pharmacol. 2010, 10, 782–788. [Google Scholar] [CrossRef]
- Brady, O.J.; Gething, P.W.; Bhatt, S.; Messina, J.P.; Brownstein, J.S.; Hoen, A.G.; Moyes, C.L.; Farlow, A.W.; Scott, T.W.; Hay, S.I. Refining the Global Spatial Limits of Dengue Virus Transmission by Evidence-Based Consensus. PLoS Negl. Trop. Dis. 2012, 6. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Bakshi, A.S. Dengue Fever, DHF and DSS. Apollo Med. 2007, 4, 111–117. [Google Scholar] [CrossRef]
- Dick, G.W.A.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.J.J. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch. Virol. 2007, 152, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Faye, O.; Freire, C.C.M.; Iamarino, A.; Faye, O.; de Oliveira, J.V.C.; Diallo, M.; Zanotto, P.M.A.; Sall, A.A. Molecular Evolution of Zika Virus during Its Emergence in the 20th Century. PLoS Negl. Trop. Dis. 2014, 8, 36. [Google Scholar] [CrossRef] [Green Version]
- Sikka, V.; Chattu, V.K.; Popli, R.K.; Galwankar, S.C.; Kelkar, D.; Sawicki, S.G.; Stawicki, S.P.; Papadimos, T.J. The Emergence of Zika Virus as a Global Health Security Threat: A Review and a Consensus Statement of the INDUSEM Joint working Group (JWG). J. Glob. Infect. Dis. 2018, 8, 3–15. [Google Scholar] [CrossRef]
- Oliveira Melo, A.S.; Malinger, G.; Ximenes, R.; Szejnfeld, P.O.; Alves Sampaio, S.; Bispo De Filippis, A.M. Zika virus intrauterine infection causes fetal brain abnormality and microcephaly: Tip of the iceberg? Ultrasound Obstet. Gynecol. 2016, 47, 6–7. [Google Scholar] [CrossRef]
- World Health Organization. WHO Malaria Report 2019; World Health Organization: Geneva, Switzerland, 2019; ISBN 9789241565721. [Google Scholar]
- Aly, A.S.I.; Vaughan, A.M.; Kappe, S.H.I. Malaria Parasite Development in the Mosquito and Infection of the Mammalian Host. Annu. Rev. Microbiol. 2009, 63, 195–221. [Google Scholar] [CrossRef] [Green Version]
- Scotland’s National Health Information Service Malaria. Available online: https://www.nhsinform.scot/illnesses-and-conditions/infections-and-poisoning/malaria#symptoms-of-malaria (accessed on 15 August 2020).
- Sager, G.; Gabaglio, S.; Sztul, E.; Belov, G.A. Role of host cell secretory machinery in zika virus life cycle. Viruses 2018, 10, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampieri, L.; Di Giusto, P.; Alvarez, C. CREB3 Transcription Factors: ER-Golgi Stress Transducers as Hubs for Cellular Homeostasis. Front. Cell Dev. Biol. 2019, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Moni, M.A.; Lio, P. Genetic Profiling and Comorbidities of Zika Infection. J. Infect. Dis. 2017, 216, 703–712. [Google Scholar] [CrossRef]
- Malaguarnera, L.; Musumeci, S. The immune response to Plasmodium falciparum malaria. Lancet Infect. Dis. 2002, 2, 472–478. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Sha, W.; Shulaev, V.; Stins, M.F.; Sullivan, D.J. Plasmodium falciparum-infected erythrocytes induce NF-κB regulated inflammatory pathways in human cerebral endothelium. Blood 2009, 114, 4243–4252. [Google Scholar] [CrossRef] [Green Version]
- Belachew, E.B. Immune Response and Evasion Mechanisms of Plasmodium falciparum Parasites. J. Immunol. Res. 2018, 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clayton, A.M.; Dong, Y.; Dimopoulos, G. The anopheles innate immune system in the defense against malaria infection. J. Innate Immun. 2014, 6, 169–181. [Google Scholar] [CrossRef]
- Souza-Neto, J.A.; Sim, S.; Dimopoulos, G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. USA 2009, 106, 17841–17846. [Google Scholar] [CrossRef] [Green Version]
- Werling, K.; Shaw, W.R.; Itoe, M.A.; Westervelt, K.A.; Marcenac, P.; Paton, D.G.; Peng, D.; Singh, N.; Smidler, A.L.; South, A.; et al. Steroid Hormone Function Controls Non-competitive Plasmodium Development in Anopheles. Cell 2019, 177, 315–325.e14. [Google Scholar] [CrossRef] [Green Version]
- Hyman, A.A.; Weber, C.A.; Jülicher, F. Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 2014, 30, 39–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitrea, D.M.; Kriwacki, R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016, 14, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef] [Green Version]
- Itakura, A.K.; Futia, R.A.; Jarosz, D.F. It Pays To Be in Phase. Biochemistry 2018, 57, 2520–2529. [Google Scholar] [CrossRef] [PubMed]
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Protein intrinsic disorder-based liquid–liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv. Colloid Interface Sci. 2017, 239, 97–114. [Google Scholar] [CrossRef]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef] [Green Version]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Posey, A.E.; Holehouse, A.S.; Pappu, R.V. Phase Separation of Intrinsically Disordered Proteins. Methods Enzymol. 2018, 611, 1–30. [Google Scholar] [CrossRef]
- Owen, I.; Shewmaker, F. The Role of Post-Translational Modifications in the Phase Transitions of Intrinsically Disordered Proteins. Int. J. Mol. Sci. 2019, 20, 5501. [Google Scholar] [CrossRef] [Green Version]
- Nair, S.J.; Yang, L.; Meluzzi, D.; Oh, S.; Yang, F.; Friedman, M.J.; Wang, S.; Suter, T.; Alshareedah, I.; Gamliel, A.; et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat. Struct. Mol. Biol. 2019, 26, 193–203. [Google Scholar] [CrossRef]
- Courchaine, E.M.; Lu, A.; Neugebauer, K.M. Droplet organelles? EMBO J. 2016, 35, 1603–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.P.; Lloyd, R.E. Regulation of stress granules in virus systems. Trends Microbiol. 2012, 20, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Wang, L.; Qin, Z.; Wang, J.; Zheng, X.; Wei, L.; Zhang, X.; Zhang, X.; Liu, C.; Li, Z.; et al. Phase Separation of Epstein-Barr Virus EBNA2 and Its Coactivator EBNALP Controls Gene Expression. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Perdikari, T.M.; Murthy, A.C.; Ryan, V.H.; Watters, S.; Naik, M.T.; Fawzi, N.L. SARS-CoV-2 nucleocapsid protein undergoes liquid-liquid phase separation stimulated by RNA and partitions into phases of human ribonucleoproteins. BioRxiv 2020. [Google Scholar] [CrossRef]
- Cubuk, J.; Alston, J.J.; Incicco, J.J.; Singh, S.; Stuchell-Brereton, M.D.; Ward, M.D.; Zimmerman, M.I.; Vithani, N.; Griffith, D.; Wagoner, J.A.; et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. BioRxiv 2020. [Google Scholar] [CrossRef]
- Alenquer, M.; Vale-Costa, S.; Etibor, T.A.; Ferreira, F.; Sousa, A.L.; Amorim, M.J. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat. Commun. 2019, 10, 1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monette, A.; Niu, M.; Chen, L.; Rao, S.; Gorelick, R.J.; Mouland, A.J. Pan-retroviral Nucleocapsid-Mediated Phase Separation Regulates Genomic RNA Positioning and Trafficking. Cell Rep. 2020, 31, 107520. [Google Scholar] [CrossRef]
- Heinrich, B.S.; Maliga, Z.; Stein, D.A.; Hyman, A.A.; Whelan, S.P.J. Phase Transitions Drive the Formation of Vesicular Stomatitis Virus Replication Compartments. mBio 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Sawyer, I.A.; Bartek, J.; Dundr, M. Phase separated microenvironments inside the cell nucleus are linked to disease and regulate epigenetic state, transcription and RNA processing. Semin. Cell Dev. Biol. 2019, 90, 94–103. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef] [Green Version]
- Tarczewska, A.; Greb-Markiewicz, B. The Significance of the Intrinsically Disordered Regions for the Functions of the bHLH Transcription Factors. Int. J. Mol. Sci. 2019, 20, 5306. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Wu, T.; Gutman, O.; Lu, H.; Zhou, Q.; Henis, Y.I.; Luo, K. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat. Cell Biol. 2020, 22, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, J.J.; Otero, J.H.; Scott, D.C.; Szulc, E.; Martin, E.W.; Sabri, N.; Granata, D.; Marzahn, M.R.; Lindorff-Larsen, K.; Salvatella, X.; et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell 2018, 72, 19–36.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.; Wang, C.; Deng, Y.; Yu, L.; Huang, H. Destruction of Full-Length Androgen Receptor by Wild-Type SPOP, but Not Prostate-Cancer-Associated Mutants. Cell Rep. 2014, 6, 657–669. [Google Scholar] [CrossRef] [Green Version]
- Brangwynne, C.P.; Tompa, P.; Pappu, R.V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015, 11, 899–904. [Google Scholar] [CrossRef]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef] [Green Version]
- Quiroz, F.G.; Chilkoti, A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 2015, 14, 1164–1171. [Google Scholar] [CrossRef]
- Stortz, M.; Presman, D.M.; Bruno, L.; Annibale, P.; Dansey, M.V.; Burton, G.; Gratton, E.; Pecci, A.; Levi, V. Mapping the Dynamics of the Glucocorticoid Receptor within the Nuclear Landscape. Sci. Rep. 2017. [Google Scholar] [CrossRef] [Green Version]
- Stortz, M.; Pecci, A.; Presman, D.M.; Levi, V. Unraveling the molecular interactions involved in phase separation of glucocorticoid receptor. BMC Biol. 2020, 18, 59. [Google Scholar] [CrossRef]
- Dao, T.P.; Kolaitis, R.-M.; Kim, H.J.; O’Donovan, K.; Martyniak, B.; Colicino, E.; Hehnly, H.; Taylor, J.P.; Castañeda, C.A. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell 2018, 69, 965–978.e6. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.R.; Skafar, D.F. Modulation of nuclear receptor activity by the F domain. Mol. Cell. Endocrinol. 2015, 418, 298–305. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Bianchetti, L.; Wassmer, B.; Defosset, A.; Smertina, A.; Tiberti, M.L.; Stote, R.H.; Dejaegere, A. Alternative dimerization interfaces in the glucocorticoid receptor-α ligand binding domain. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1810–1825. [Google Scholar] [CrossRef]
- Arao, Y.; Korach, K.S. The F domain of estrogen receptor α is involved in species-specific, tamoxifen-mediated transactivation. J. Biol. Chem. 2018, 293, 8495–8507. [Google Scholar] [CrossRef] [Green Version]
- Guzman, M.G.; Halstead, S.B.; Artsob, H.; Buchy, P.; Farrar, J.; Nathan, M.B.; Pelegrino, J.L.; Simmons, C.; Yoksan, S. Dengue: A continuing global threat Europe PMC Funders Author Manuscripts. Nat. Rev. Microbiol. 2010, 8, S7–S16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchette, N.J.; Garcia, R.; Rudnick, A. Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am. J. Trop. Med. Hyg. 1969, 18, 411–415. [Google Scholar] [CrossRef]
- Barrett, A.D.T.; Higgs, S. Yellow fever: A disease that has yet to be conquered. Annu. Rev. Entomol. 2007, 52, 209–229. [Google Scholar] [CrossRef] [Green Version]
- Tsetsarkin, K.A.; Chen, R.; Weaver, S.C. Interspecies transmission and chikungunya virus emergence. Curr. Opin. Virol. 2016, 16, 143–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raikhel, A.S.; Kokoza, V.; Zhu, J.; Martin, D.; Wang, S.F.; Li, C.; Sun, G.; Ahmed, A.; Dittmer, N.; Attardo, G. Molecular biology of mosquito vitellogenesis: From basic studies to genetic engineering of antipathogen immunity. Insect Biochem. Mol. Biol. 2002, 32, 1275–1286. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Ranieri-Raggi, M.; Moir, A.; Raggi, A. The Role of Histidine-Proline-Rich Glycoprotein as Zinc Chaperone for Skeletal Muscle AMP Deaminase. Biomolecules 2014, 4, 474–497. [Google Scholar] [CrossRef] [Green Version]
- Adzhubei, A.A.; Sternberg, M.J.E.; Makarov, A.A. Polyproline-II helix in proteins: Structure and function. J. Mol. Biol. 2013, 425, 2100–2132. [Google Scholar] [CrossRef] [PubMed]
- Kay, B.K.; Williamson, M.P.; Sudol, M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000, 14, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.M.; Hsu, V.L. The Extended Left-Handed Helix: A Simple Nucleic Acid-Binding Motif. Proteins Struct. Funct. Genet. 2004, 55, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Siligardi, G.; Drake, A.F. The importance of extended conformations and, in particular, the PII conformation for the molecular recognition of peptides. Biopolymers 1995, 37, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Vernon, R.M.; Chong, P.A.; Tsang, B.; Kim, T.H.; Bah, A.; Farber, P.; Lin, H.; Forman-Kay, J.D. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 2018, 7. [Google Scholar] [CrossRef]
- Horvath, A.; Miskei, M.; Ambrus, V.; Vendruscolo, M.; Fuxreiter, M. Sequence-based prediction of protein binding mode landscapes. PLoS Comput. Biol. 2020, 16, e1007864. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, K.; Song, D.; Jung, Y. Behavior control of membrane-less protein liquid condensates with metal ion-induced phase separation. Nat. Commun. 2020, 11, 5554. [Google Scholar] [CrossRef]
- Monette, A.; Mouland, A.J. Zinc and Copper Ions Differentially Regulate Prion-Like Phase Separation Dynamics of Pan-Virus Nucleocapsid Biomolecular Condensates. Viruses 2020, 12, 1179. [Google Scholar] [CrossRef]
- Rao, A.K.; Ziegler, Y.S.; McLeod, I.X.; Yates, J.R.; Nardulli, A.M. Effects of Cu/Zn superoxide dismutase on estrogen responsiveness and oxidative stress in human breast cancer cells. Mol. Endocrinol. 2008, 22, 1113–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FDA First FDA-Approved Vaccine for the Prevention of Dengue Disease in Endemic Regions. Available online: https://www.fda.gov/news-events/press-announcements/first-fda-approved-vaccine-prevention-dengue-disease-endemic-regions (accessed on 1 May 2020).
- Alonso, P.L.; Sacarlal, J.; Aponte, J.J.; Leach, A.; Macete, E.; Milman, J.; Mandomando, I.; Spiessens, B.; Guinovart, C.; Espasa, M.; et al. Efficacy of the RTS,S/AS02A vaccine against Plasmodium falciparum infection and disease in young African children: Randomised controlled trial. Lancet 2004, 364, 1411–1420. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention How to Reduce Malaria’s Impact. Available online: https://www.cdc.gov/malaria/malaria_worldwide/reduction/vaccine.html (accessed on 1 May 2020).
- Fung, H.Y.J.; Birol, M.; Rhoades, E. IDPs in macromolecular complexes: The roles of multivalent interactions in diverse assemblies. Curr. Opin. Struct. Biol. 2018, 49, 36–43. [Google Scholar] [CrossRef]
- Ekoka, E.; Maharaj, S.; Nardini, L.; Dahan-Moss, Y.; Koekemoer, L.L. 20-Hydroxyecdysone (20E) signaling as a promising target for the chemical control of malaria vectors. Parasites Vectors 2021, 14, 86. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Więch, A.; Tarczewska, A.; Ożyhar, A.; Orłowski, M. Metal Ions Induce Liquid Condensate Formation by the F Domain of Aedes aegypti Ecdysteroid Receptor. New Perspectives of Nuclear Receptor Studies. Cells 2021, 10, 571. https://doi.org/10.3390/cells10030571
Więch A, Tarczewska A, Ożyhar A, Orłowski M. Metal Ions Induce Liquid Condensate Formation by the F Domain of Aedes aegypti Ecdysteroid Receptor. New Perspectives of Nuclear Receptor Studies. Cells. 2021; 10(3):571. https://doi.org/10.3390/cells10030571
Chicago/Turabian StyleWięch, Anna, Aneta Tarczewska, Andrzej Ożyhar, and Marek Orłowski. 2021. "Metal Ions Induce Liquid Condensate Formation by the F Domain of Aedes aegypti Ecdysteroid Receptor. New Perspectives of Nuclear Receptor Studies" Cells 10, no. 3: 571. https://doi.org/10.3390/cells10030571
APA StyleWięch, A., Tarczewska, A., Ożyhar, A., & Orłowski, M. (2021). Metal Ions Induce Liquid Condensate Formation by the F Domain of Aedes aegypti Ecdysteroid Receptor. New Perspectives of Nuclear Receptor Studies. Cells, 10(3), 571. https://doi.org/10.3390/cells10030571