Enhanced Intestinal TGF-β/SMAD-Dependent Signaling in Simian Immunodeficiency Virus Infected Rhesus Macaques
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals, Ethics Statement, and Tissue Sampling
2.2. Plasma Viral Load Quantification
2.3. Isolation and Enrichment of Lymphocytes from Intestine
2.4. Immunohistochemistry Staining
2.5. Flow Cytometry Analysis
2.6. Immunofluorescence Assay
2.7. Quantification of SMAD3 and SMAD7 Gene Expression in Jejunum Tissues
2.8. Statistical Analysis and Graphical Presentation
3. Results
3.1. Terminal Plasma Viral Loads in SIV Infected Macaques
3.2. TGF-β Production Increases in the Jejunum of SIV-Infected Macaques
3.3. Characterization of TGF-β Expressing Cell Populations in LPLs
3.4. TGF-β Receptor II Expression Remains Unaffected during SIV Infection
3.5. An Increased Level of pSMAD2-3 Complex Suggests Persistent Activation of Signaling
3.6. Upregulation of SMAD3, A Positive TGF-β/SMAD Signaling Regulator Is Associated with Disease Progression
3.7. Downregulation of SMAD7 Expression Denotes the Failure of Negative Feedback Mechanism
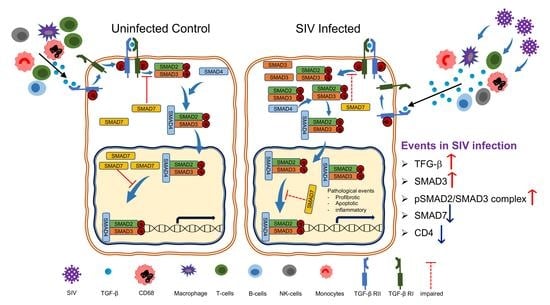
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pahar, B.; Pan, D.; Lala, W.; Kenway-Lynch, C.S.; Das, A. Transforming growth factor-β1 regulated phosphorylated AKT and interferon gamma expressions are associated with epithelial cell survival in rhesus macaque colon explants. Clin. Immunol. 2015, 158, 8–18. [Google Scholar] [CrossRef] [Green Version]
- Blobe, G.C.; Schiemann, W.P.; Lodish, H.F. Role of transforming growth factor β in human disease. N. Engl. J. Med. 2000, 342, 1350–1358. [Google Scholar] [CrossRef]
- Kubiczkova, L.; Sedlarikova, L.; Hajek, R.; Sevcikova, S. TGF-β—An excellent servant but a bad master. J. Transl. Med. 2012, 10, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakefield, L.M.; Roberts, A.B. TGF-β signaling: Positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 2002, 12, 22–29. [Google Scholar] [CrossRef]
- Derynck, R.; Budi, E.H. Specificity, versatility, and control of TGF-β family signaling. Sci. Signal. 2019, 12. [Google Scholar] [CrossRef] [Green Version]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-β and the TGF-β Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef] [Green Version]
- Attisano, L.; Lee-Hoeflich, S.T. The Smads. Genome Biol. 2001, 2. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Chen, Y.G. TGF-β Signaling from Receptors to Smads. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Do, T.V.; Kubba, L.A.; Du, H.; Sturgis, C.D.; Woodruff, T.K. Transforming growth factor-β1, transforming growth factor-β2, and transforming growth factor-β3 enhance ovarian cancer metastatic potential by inducing a Smad3-dependent epithelial-to-mesenchymal transition. Mol. Cancer Res. 2008, 6, 695–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.H.; Chen, D.Q.; Wang, Y.N.; Feng, Y.L.; Cao, G.; Vaziri, N.D.; Zhao, Y.Y. New insights into TGF-β/Smad signaling in tissue fibrosis. Chem. Biol. Interact. 2018, 292, 76–83. [Google Scholar] [CrossRef] [Green Version]
- Quezada, M.; Wang, J.; Hoang, V.; McGee, E.A. Smad7 is a transforming growth factor-β-inducible mediator of apoptosis in granulosa cells. Fertil. Steril. 2012, 97, 1452–1459.e1–6. [Google Scholar] [CrossRef]
- Lonn, P.; Moren, A.; Raja, E.; Dahl, M.; Moustakas, A. Regulating the stability of TGFβ receptors and Smads. Cell Res. 2009, 19, 21–35. [Google Scholar] [CrossRef]
- Yan, X.; Liao, H.; Cheng, M.; Shi, X.; Lin, X.; Feng, X.H.; Chen, Y.G. Smad7 Protein Interacts with Receptor-regulated Smads (R-Smads) to Inhibit Transforming Growth Factor-β (TGF-β)/Smad Signaling. J. Biol. Chem. 2016, 291, 382–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Fei, T.; Zhang, L.; Zhang, R.; Chen, F.; Ning, Y.; Han, Y.; Feng, X.H.; Meng, A.; Chen, Y.G. Smad7 antagonizes transforming growth factor β signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol. Cell. Biol. 2007, 27, 4488–4499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.E. Non-Smad Signaling Pathways of the TGF-β Family. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Maina, E.K.; Abana, C.Z.; Bukusi, E.A.; Sedegah, M.; Lartey, M.; Ampofo, W.K. Plasma concentrations of transforming growth factor β 1 in non-progressive HIV-1 infection correlates with markers of disease progression. Cytokine 2016, 81, 109–116. [Google Scholar] [CrossRef] [Green Version]
- Cumont, M.C.; Monceaux, V.; Viollet, L.; Lay, S.; Parker, R.; Hurtrel, B.; Estaquier, J. TGF-β in intestinal lymphoid organs contributes to the death of armed effector CD8 T cells and is associated with the absence of virus containment in rhesus macaques infected with the simian immunodeficiency virus. Cell Death Differ. 2007, 14, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guan, E.; Roderiquez, G.; Norcross, M.A. Synergistic induction of apoptosis in primary CD4(+) T cells by macrophage-tropic HIV-1 and TGF-β1. J. Immunol. 2001, 167, 3360–3366. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Smith, A.J.; Wietgrefe, S.W.; Southern, P.J.; Schacker, T.W.; Reilly, C.S.; Estes, J.D.; Burton, G.F.; Silvestri, G.; Lifson, J.D.; et al. Cumulative mechanisms of lymphoid tissue fibrosis and T cell depletion in HIV-1 and SIV infections. J. Clin. Investig. 2011, 121, 998–1008. [Google Scholar] [CrossRef] [PubMed]
- Theron, A.J.; Anderson, R.; Rossouw, T.M.; Steel, H.C. The Role of Transforming Growth Factor Beta-1 in the Progression of HIV/AIDS and Development of Non-AIDS-Defining Fibrotic Disorders. Front. Immunol. 2017, 8, 1461. [Google Scholar] [CrossRef]
- Tang, J.; Gifford, C.C.; Samarakoon, R.; Higgins, P.J. Deregulation of Negative Controls on TGF-β1 Signaling in Tumor Progression. Cancers 2018, 10, 159. [Google Scholar] [CrossRef] [Green Version]
- Kashima, R.; Hata, A. The role of TGF-β superfamily signaling in neurological disorders. Acta Biochim. Biophys. Sin. 2018, 50, 106–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, F.; Jain, M.K.; Feinberg, M.W.; Sibinga, N.E.; Pellacani, A.; Wiesel, P.; Chin, M.T.; Topper, J.N.; Perrella, M.A.; Lee, M.E. Transforming growth factor-β 1 inhibition of macrophage activation is mediated via Smad3. J. Biol. Chem. 2000, 275, 36653–36658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimura, A.; Wakabayashi, Y.; Mori, T. Cellular and molecular basis for the regulation of inflammation by TGF-β. J. Biochem. 2010, 147, 781–792. [Google Scholar] [CrossRef]
- Misra, A.; Thippeshappa, R.; Kimata, J.T. Macaques as model hosts for studies of HIV-1 infection. Front. Microbiol. 2013, 4, 176. [Google Scholar] [CrossRef] [Green Version]
- Pahar, B.; Kuebler, D.; Rasmussen, T.; Wang, X.; Srivastav, S.K.; Das, A.; Veazey, R.S. Quantification of Viral RNA and DNA Positive Cells in Tissues From Simian Immunodeficiency Virus/Simian Human Immunodeficiency Virus Infected Controller and Progressor Rhesus Macaques. Front. Microbiol. 2019, 10, 2933. [Google Scholar] [CrossRef] [PubMed]
- Pahar, B.; Kenway-Lynch, C.S.; Marx, P.; Srivastav, S.K.; LaBranche, C.; Montefiori, D.C.; Das, A. Breadth and magnitude of antigen-specific antibody responses in the control of plasma viremia in simian immunodeficiency virus infected macaques. Virol. J. 2016, 13, 200. [Google Scholar] [CrossRef] [Green Version]
- Pan, D.; Kenway-Lynch, C.S.; Lala, W.; Veazey, R.S.; Lackner, A.A.; Das, A.; Pahar, B. Lack of interleukin-10-mediated anti-inflammatory signals and upregulated interferon gamma production are linked to increased intestinal epithelial cell apoptosis in pathogenic simian immunodeficiency virus infection. J. Virol. 2014, 88, 13015–13028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenway-Lynch, C.S.; Das, A.; Lackner, A.A.; Pahar, B. Cytokine/Chemokine responses in activated CD4+ and CD8+ T cells isolated from peripheral blood, bone marrow, and axillary lymph nodes during acute simian immunodeficiency virus infection. J. Virol. 2014, 88, 9442–9457. [Google Scholar] [CrossRef] [Green Version]
- Pahar, B.; Lackner, A.A.; Veazey, R.S. Intestinal double-positive CD4+CD8+ T cells are highly activated memory cells with an increased capacity to produce cytokines. Eur. J. Immunol. 2006, 36, 583–592. [Google Scholar] [CrossRef]
- Pan, D.; Das, A.; Liu, D.; Veazey, R.S.; Pahar, B. Isolation and characterization of intestinal epithelial cells from normal and SIV-infected rhesus macaques. PLoS ONE 2012, 7, e30247. [Google Scholar] [CrossRef] [Green Version]
- Pan, D.; Das, A.; Srivastav, S.K.; Traina-Dorge, V.; Didier, P.J.; Pahar, B. Lack of T-cell-mediated IL-2 and TNFalpha production is linked to decreased CD58 expression in intestinal tissue during acute simian immunodeficiency virus infection. J. Gen. Virol. 2019, 100, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Franze, E.; Caruso, R.; Stolfi, C.; Sarra, M.; Cupi, M.L.; Caprioli, F.; Monteleone, I.; Zorzi, F.; De Nitto, D.; Colantoni, A.; et al. Lesional accumulation of CD163-expressing cells in the gut of patients with inflammatory bowel disease. PLoS ONE 2013, 8, e69839. [Google Scholar] [CrossRef]
- Li, H.; Reeves, R.K. Functional perturbation of classical natural killer and innate lymphoid cells in the oral mucosa during SIV infection. Front. Immunol. 2012, 3, 417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiercinska-Drapalo, A.; Flisiak, R.; Jaroszewicz, J.; Prokopowicz, D. Increased plasma transforming growth factor-β1 is associated with disease progression in HIV-1-infected patients. Viral Immunol. 2004, 17, 109–113. [Google Scholar] [CrossRef]
- Asmuth, D.M.; Pinchuk, I.V.; Wu, J.; Vargas, G.; Chen, X.; Mann, S.; Albanese, A.; Ma, Z.M.; Saroufeem, R.; Melcher, G.P.; et al. Role of intestinal myofibroblasts in HIV-associated intestinal collagen deposition and immune reconstitution following combination antiretroviral therapy. AIDS 2015, 29, 877–888. [Google Scholar] [CrossRef] [Green Version]
- Kornfeld, C.; Ploquin, M.J.; Pandrea, I.; Faye, A.; Onanga, R.; Apetrei, C.; Poaty-Mavoungou, V.; Rouquet, P.; Estaquier, J.; Mortara, L.; et al. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J. Clin. Investig. 2005, 115, 1082–1091. [Google Scholar] [CrossRef]
- Gottfried, E.; Kunz-Schughart, L.A.; Weber, A.; Rehli, M.; Peuker, A.; Muller, A.; Kastenberger, M.; Brockhoff, G.; Andreesen, R.; Kreutz, M. Expression of CD68 in non-myeloid cell types. Scand. J. Immunol. 2008, 67, 453–463. [Google Scholar] [CrossRef]
- Amanzada, A.; Malik, I.A.; Blaschke, M.; Khan, S.; Rahman, H.; Ramadori, G.; Moriconi, F. Identification of CD68(+) neutrophil granulocytes in in vitro model of acute inflammation and inflammatory bowel disease. Int. J. Clin. Exp. Pathol. 2013, 6, 561–570. [Google Scholar] [CrossRef] [Green Version]
- Mowat, A.M.; Bain, C.C. Mucosal macrophages in intestinal homeostasis and inflammation. J. Innate Immun. 2011, 3, 550–564. [Google Scholar] [CrossRef] [PubMed]
- Vander Ark, A.; Cao, J.; Li, X. TGF-β receptors: In and beyond TGF-β signaling. Cell. Signal. 2018, 52, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Groppe, J.; Hinck, C.S.; Samavarchi-Tehrani, P.; Zubieta, C.; Schuermann, J.P.; Taylor, A.B.; Schwarz, P.M.; Wrana, J.L.; Hinck, A.P. Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell 2008, 29, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.; Padidam, M.; Cress, D.; Grady, W.M. TGF-β receptor levels regulate the specificity of signaling pathway activation and biological effects of TGF-β. Biochim. Biophys. Acta 2009, 1793, 1165–1173. [Google Scholar] [CrossRef] [Green Version]
- Duan, D.; Derynck, R. Transforming growth factor-β (TGF-β)-induced up-regulation of TGF-β receptors at the cell surface amplifies the TGF-β response. J. Biol. Chem. 2019, 294, 8490–8504. [Google Scholar] [CrossRef]
- Hougaard, S.; Norgaard, P.; Abrahamsen, N.; Moses, H.L.; Spang-Thomsen, M.; Skovgaard Poulsen, H. Inactivation of the transforming growth factor β type II receptor in human small cell lung cancer cell lines. Br. J. Cancer 1999, 79, 1005–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Derynck, R. Essential role of TGF-β signaling in glucose-induced cell hypertrophy. Dev. Cell 2009, 17, 35–48. [Google Scholar] [CrossRef] [Green Version]
- Massague, J. How cells read TGF-β signals. Nat. Rev. Mol. Cell Biol. 2000, 1, 169–178. [Google Scholar] [CrossRef]
- Dalvi, P.; Sharma, H.; Konstantinova, T.; Sanderson, M.; Brien-Ladner, A.O.; Dhillon, N.K. Hyperactive TGF-β Signaling in Smooth Muscle Cells Exposed to HIV-protein(s) and Cocaine: Role in Pulmonary Vasculopathy. Sci. Rep. 2017, 7, 10433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flanders, K.C. Smad3 as a mediator of the fibrotic response. Int. J. Exp. Pathol. 2004, 85, 47–64. [Google Scholar] [CrossRef]
- Meng, X.M.; Tang, P.M.; Li, J.; Lan, H.Y. TGF-β/Smad signaling in renal fibrosis. Front. Physiol. 2015, 6, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, X.; Liu, Z.; Chen, Y. Regulation of TGF-β signaling by Smad7. Acta Biochim. Biophys. Sin. 2009, 41, 263–272. [Google Scholar] [CrossRef] [Green Version]
- Brodin, G.; Ahgren, A.; ten Dijke, P.; Heldin, C.H.; Heuchel, R. Efficient TGF-β induction of the Smad7 gene requires cooperation between AP-1, Sp1, and Smad proteins on the mouse Smad7 promoter. J. Biol. Chem. 2000, 275, 29023–29030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakao, A.; Afrakhte, M.; Moren, A.; Nakayama, T.; Christian, J.L.; Heuchel, R.; Itoh, S.; Kawabata, M.; Heldin, N.E.; Heldin, C.H.; et al. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature 1997, 389, 631–635. [Google Scholar] [CrossRef]
- Luwor, R.B.; Baradaran, B.; Taylor, L.E.; Iaria, J.; Nheu, T.V.; Amiry, N.; Hovens, C.M.; Wang, B.; Kaye, A.H.; Zhu, H.J. Targeting Stat3 and Smad7 to restore TGF-β cytostatic regulation of tumor cells in vitro and in vivo. Oncogene 2013, 32, 2433–2441. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Category | Animal Number | Age (Year) | Sex a | Virus | Days of Infection | Dosage (TCID50) | Route b | Terminal Plasma Viral Load (RNA Copies/mL) |
|---|---|---|---|---|---|---|---|---|
| Uninfected Control | AG71 | 11.1 | F | Nil | - | - | - | - |
| DJ78 | 8.1 | F | Nil | - | - | - | - | |
| EV39 | 6.2 | M | Nil | - | - | - | - | |
| FK25 | 5.8 | M | Nil | - | - | - | - | |
| GI92 | 4.2 | M | Nil | - | - | - | - | |
| GN70 | 10.1 | F | Nil | - | - | - | - | |
| GN74 | 13.3 | F | Nil | - | - | - | - | |
| GT20 | 4.6 | M | Nil | - | - | - | - | |
| HG11 | 7.1 | M | Nil | - | - | - | - | |
| HG64 | 3.0 | M | Nil | - | - | - | - | |
| JB65 | 7.6 | M | Nil | - | - | - | - | |
| JG16 | 4.3 | M | Nil | - | - | - | - | |
| JM76 | 6.3 | M | Nil | - | - | - | - | |
| Acute SIV | AV91 | 14.1 | M | SIVMAC251 | 10 | 500 | IV | 157,190,000 |
| BA57 | 14 | F | SIVMAC251 | 8 | 500 | IV | 14,288,200 | |
| BN37 | 2.5 | M | SIVMAC251 | 21 | 100 | IV | 340,000 | |
| CF65 | 12.3 | F | SIVMAC251 | 21 | 500 | IVAG | 10,100,000 | |
| EK98 | 8.7 | F | SIVMAC251 | 21 | 500 | IVAG | 26,800,000 | |
| EM64 | 8.9 | F | SIVMAC251 | 21 | 500 | IVAG | 3,840,000 | |
| FT35 | 6.7 | F | SIVMAC251 | 21 | 500 | IVAG | 3,540,000 | |
| GI28 | 5.9 | F | SIVMAC251 | 21 | 500 | IVAG | 5,830,000 | |
| HI53 | 6.6 | F | SIVMAC251 | 8 | 100 | IV | 3,555,700 | |
| HN29 | 12.6 | F | SIVMAC251 | 10 | 100 | IV | 110,000,000 | |
| M992 | 16 | F | SIVMAC251 | 13 | 500 | IV | 34,949,800 | |
| Chronic SIV | BC35 | 11.9 | F | SIVMAC251 | 422 | 300 | IVAG | 428,298 |
| BD03 | 12.8 | F | SIVMAC251 | 167 | 500 | IVAG | 14,788,890 | |
| CL86 | 11.1 | F | SIVMAC251 | 281 | 500 | IVAG | 972,889 | |
| DE50 | 9.7 | F | SIVMAC251 | 150 | 500 | IVAG | 304,000 | |
| DR59 | 6.3 | F | SIVMAC251 | 250 | 1000 | IVAG | 288,441 | |
| EB09 | 6.3 | F | SIVMAC251 | 250 | 100 | IVAG | 750,720 | |
| EJ26 | 6.2 | F | SIVMAC251 | 309 | 100 | IV | 397,806 | |
| FK88 | 6.5 | F | SIVMAC251 | 226 | 500 | IVAG | 2,314,583 | |
| GN91 | 4.9 | F | SIVMAC251 | 401 | 500 | IVAG | 4190 | |
| HG58 | 9.1 | F | SIVMAC251 | 288 | 300 | IVAG | 7074 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boby, N.; Ransom, A.; Pace, B.T.; Williams, K.M.; Mabee, C.; Das, A.; Srivastav, S.K.; Porter, E.; Pahar, B. Enhanced Intestinal TGF-β/SMAD-Dependent Signaling in Simian Immunodeficiency Virus Infected Rhesus Macaques. Cells 2021, 10, 806. https://doi.org/10.3390/cells10040806
Boby N, Ransom A, Pace BT, Williams KM, Mabee C, Das A, Srivastav SK, Porter E, Pahar B. Enhanced Intestinal TGF-β/SMAD-Dependent Signaling in Simian Immunodeficiency Virus Infected Rhesus Macaques. Cells. 2021; 10(4):806. https://doi.org/10.3390/cells10040806
Chicago/Turabian StyleBoby, Nongthombam, Alyssa Ransom, Barcley T. Pace, Kelsey M. Williams, Christopher Mabee, Arpita Das, Sudesh K. Srivastav, Edith Porter, and Bapi Pahar. 2021. "Enhanced Intestinal TGF-β/SMAD-Dependent Signaling in Simian Immunodeficiency Virus Infected Rhesus Macaques" Cells 10, no. 4: 806. https://doi.org/10.3390/cells10040806
APA StyleBoby, N., Ransom, A., Pace, B. T., Williams, K. M., Mabee, C., Das, A., Srivastav, S. K., Porter, E., & Pahar, B. (2021). Enhanced Intestinal TGF-β/SMAD-Dependent Signaling in Simian Immunodeficiency Virus Infected Rhesus Macaques. Cells, 10(4), 806. https://doi.org/10.3390/cells10040806