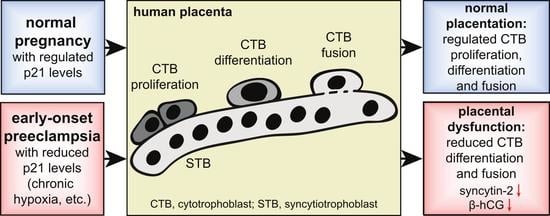
Functional Analysis of p21Cip1/CDKN1A and Its Family Members in Trophoblastic Cells of the Placenta and Its Roles in Preeclampsia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Placental Tissue Collection
2.2. Formation of Trophoblast Organoids from Human First Trimester Placental Tissue
2.3. Preparation of Organoids and Placental Tissues for IHC-IF
2.4. Isolation of Primary Villous Cytotrophoblasts from Human First Trimester Placenta
2.5. Isolation of Placental Mesenchymal Stem/Stromal Cells from First Trimester Placental Tissue
2.6. Isolation of Primary Villous Cytotrophoblasts from Human Term Placental Tissue
2.7. Immunohistochemistry of Placental Tissue
2.8. RNA Extraction and Real-Time Quantitative PCR
2.9. Cell Culture, Transfection and Treatment
2.10. Western Blot Analysis
2.11. Immunofluorescence Staining
2.12. Luciferase Assay
2.13. Microarray Analysis
2.14. Statistical Analysis
3. Results
3.1. Cell Cycle Regulators Are Affected by the Delivery Mode, and Specifically Expressed in Trophoblast Organoids and Placental Tissues
3.2. Cell Cycle Regulators Are Highly Expressed during the First Trimester of Gestation
3.3. p21 Expression Is Reduced in fCTBs of Early-Onset PE Placental Samples
3.4. Reduced p21 Protein Expression in Early-Onset PE with HELLP Syndrome
3.5. p21 Expression Is Decreased in Trophoblastic Cell Lines and in Isolated Primary Cytotrophoblasts under Hypoxic Conditions
3.6. Knockdown of p21 Impairs the Fusion Ability of Trophoblastic BeWo Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steegers, E.A.; von Dadelszen, P.; Duvekot, J.J.; Pijnenborg, R. Pre-eclampsia. Lancet 2010, 376, 631–644. [Google Scholar] [CrossRef]
- Phipps, E.A.; Thadhani, R.; Benzing, T.; Karumanchi, S.A. Pre-eclampsia: Pathogenesis, novel diagnostics and therapies. Nat Rev. Nephrol. 2019, 15, 275–289. [Google Scholar] [CrossRef]
- Roberts, J.M.; Taylor, R.N.; Musci, T.J.; Rodgers, G.M.; Hubel, C.A.; McLaughlin, M.K. Preeclampsia: An endothelial cell disorder. Am. J. Obstet. Gynecol. 1989, 161, 1200–1204. [Google Scholar] [CrossRef]
- Myatt, L.; Roberts, J.M. Preeclampsia: Syndrome or Disease? Curr. Hypertens. Rep. 2015, 17, 83. [Google Scholar] [CrossRef] [PubMed]
- Robillard, P.Y.; Dekker, G.; Chaouat, G.; Scioscia, M.; Iacobelli, S.; Hulsey, T.C. Historical evolution of ideas on eclampsia/preeclampsia: A proposed optimistic view of preeclampsia. J. Reprod. Immunol. 2017, 123, 72–77. [Google Scholar] [CrossRef]
- Phipps, E.; Prasanna, D.; Brima, W.; Jim, B. Preeclampsia: Updates in Pathogenesis, Definitions, and Guidelines. Clin. J. Am. Soc. Nephrol. 2016, 11, 1102–1113. [Google Scholar] [CrossRef] [Green Version]
- Weinstein, L. Syndrome of hemolysis, elevated liver enzymes, and low platelet count: A severe consequence of hypertension in pregnancy. Am. J. Obstet. Gynecol. 1982, 142, 159–167. [Google Scholar] [CrossRef]
- Rana, S.; Lemoine, E.; Granger, J.P.; Karumanchi, S.A. Preeclampsia: Pathophysiology, Challenges, and Perspectives. Circ. Res. 2019, 124, 1094–1112. [Google Scholar] [CrossRef] [PubMed]
- Turco, M.Y.; Moffett, A. Development of the human placenta. Development 2019, 146, dev163428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haram, K.; Mortensen, J.H.; Nagy, B. Genetic aspects of preeclampsia and the HELLP syndrome. J. Pregnancy 2014, 2014, 910751. [Google Scholar] [CrossRef] [PubMed]
- Knofler, M.; Pollheimer, J. Human placental trophoblast invasion and differentiation: A particular focus on Wnt signaling. Front Genet. 2013, 4, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coutifaris, C.; Kao, L.C.; Sehdev, H.M.; Chin, U.; Babalola, G.O.; Blaschuk, O.W.; Strauss, J.F., 3rd. E-cadherin expression during the differentiation of human trophoblasts. Development 1991, 113, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Gauster, M.; Huppertz, B. Fusion of cytotrophoblast with syncytiotrophoblast in the human placenta: Factors involved in syncytialization. J. Reprod. Med. Endocrinol. 2008, 5, 76–82. [Google Scholar]
- Aplin, J.D.; Jones, C.J.P. Cell dynamics in human villous trophoblast. Hum. Reprod. Update 2021, 27, 904–922. [Google Scholar] [CrossRef]
- Knofler, M.; Haider, S.; Saleh, L.; Pollheimer, J.; Gamage, T.; James, J. Human placenta and trophoblast development: Key molecular mechanisms and model systems. Cell Mol. Life Sci. 2019, 76, 3479–3496. [Google Scholar] [CrossRef] [Green Version]
- O’Tierney-Ginn, P.F.; Lash, G.E. Beyond pregnancy: Modulation of trophoblast invasion and its consequences for fetal growth and long-term children’s health. J. Reprod. Immunol. 2014, 104–105, 37–42. [Google Scholar] [CrossRef]
- Sanchez-Aranguren, L.C.; Prada, C.E.; Riano-Medina, C.E.; Lopez, M. Endothelial dysfunction and preeclampsia: Role of oxidative stress. Front. Physiol. 2014, 5, 372. [Google Scholar] [CrossRef] [Green Version]
- Heazell, A.E.; Lacey, H.A.; Jones, C.J.; Huppertz, B.; Baker, P.N.; Crocker, I.P. Effects of oxygen on cell turnover and expression of regulators of apoptosis in human placental trophoblast. Placenta 2008, 29, 175–186. [Google Scholar] [CrossRef]
- Huppertz, B.; Kadyrov, M.; Kingdom, J.C. Apoptosis and its role in the trophoblast. Am. J. Obstet. Gynecol. 2006, 195, 29–39. [Google Scholar] [CrossRef]
- Fisher, S.J. The placental problem: Linking abnormal cytotrophoblast differentiation to the maternal symptoms of preeclampsia. Reprod. Biol. Endocrinol. 2004, 2, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Ruijtenberg, S.; van den Heuvel, S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 2016, 15, 196–212. [Google Scholar] [CrossRef] [Green Version]
- Warfel, N.A.; El-Deiry, W.S. p21WAF1 and tumourigenesis: 20 years after. Curr. Opin. Oncol. 2013, 25, 52–58. [Google Scholar] [CrossRef]
- Kreis, N.N.; Louwen, F.; Yuan, J. Less understood issues: p21(Cip1) in mitosis and its therapeutic potential. Oncogene 2015, 34, 1758–1767. [Google Scholar] [CrossRef] [PubMed]
- Kreis, N.N.; Louwen, F.; Yuan, J. The Multifaceted p21 (Cip1/Waf1/CDKN1A) in Cell Differentiation, Migration and Cancer Therapy. Cancers 2019, 11, 1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreis, N.N.; Friemel, A.; Ritter, A.; Roth, S.; Rolle, U.; Louwen, F.; Yuan, J. Function of p21 (Cip1/Waf1/CDKN1A) in Migration and Invasion of Cancer and Trophoblastic Cells. Cancers 2019, 11, 989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheridan, M.A.; Fernando, R.C.; Gardner, L.; Hollinshead, M.S.; Burton, G.J.; Moffett, A.; Turco, M.Y. Establishment and differentiation of long-term trophoblast organoid cultures from the human placenta. Nat. Protoc. 2020, 15, 3441–3463. [Google Scholar] [CrossRef]
- Ritter, A.; Friemel, A.; Fornoff, F.; Adjan, M.; Solbach, C.; Yuan, J.; Louwen, F. Characterization of adipose-derived stem cells from subcutaneous and visceral adipose tissues and their function in breast cancer cells. Oncotarget 2015, 6, 34475–34493. [Google Scholar] [CrossRef] [Green Version]
- Turco, M.Y.; Gardner, L.; Kay, R.G.; Hamilton, R.S.; Prater, M.; Hollinshead, M.S.; McWhinnie, A.; Esposito, L.; Fernando, R.; Skelton, H.; et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 2018, 564, 263–267. [Google Scholar] [CrossRef] [Green Version]
- Haider, S.; Meinhardt, G.; Saleh, L.; Kunihs, V.; Gamperl, M.; Kaindl, U.; Ellinger, A.; Burkard, T.R.; Fiala, C.; Pollheimer, J.; et al. Self-Renewing Trophoblast Organoids Recapitulate the Developmental Program of the Early Human Placenta. Stem Cell Rep. 2018, 11, 537–551. [Google Scholar] [CrossRef] [Green Version]
- Vondra, S.; Kunihs, V.; Eberhart, T.; Eigner, K.; Bauer, R.; Haslinger, P.; Haider, S.; Windsperger, K.; Klambauer, G.; Schutz, B.; et al. Metabolism of cholesterol and progesterone is differentially regulated in primary trophoblastic subtypes and might be disturbed in recurrent miscarriages. J. Lipid Res. 2019, 60, 1922–1934. [Google Scholar] [CrossRef]
- Petroff, M.G.; Phillips, T.A.; Ka, H.; Pace, J.L.; Hunt, J.S. Isolation and culture of term human trophoblast cells. Methods Mol. Med. 2006, 121, 203–217. [Google Scholar]
- Steigman, S.A.; Fauza, D.O. Isolation of Mesenchymal Stem Cells from Amniotic Fluid and Placenta. Curr. Protoc. Stem Cell Biol. 2007, 1, 1E.2.1–1E.2.12. [Google Scholar] [CrossRef] [PubMed]
- Rodel, F.; Steinhauser, K.; Kreis, N.N.; Friemel, A.; Martin, D.; Wieland, U.; Rave-Frank, M.; Balermpas, P.; Fokas, E.; Louwen, F.; et al. Prognostic impact of RITA expression in patients with anal squamous cell carcinoma treated with chemoradiotherapy. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2018, 126, 214–221. [Google Scholar] [CrossRef]
- Detre, S.; Saclani Jotti, G.; Dowsett, M. A “quickscore” method for immunohistochemical semiquantitation: Validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol. 1995, 48, 876–878. [Google Scholar] [CrossRef] [Green Version]
- McClelland, R.A.; Finlay, P.; Walker, K.J.; Nicholson, D.; Robertson, J.F.; Blamey, R.W.; Nicholson, R.I. Automated quantitation of immunocytochemically localized estrogen receptors in human breast cancer. Cancer Res. 1990, 50, 3545–3550. [Google Scholar] [PubMed]
- Meller, M.; Vadachkoria, S.; Luthy, D.A.; Williams, M.A. Evaluation of housekeeping genes in placental comparative expression studies. Placenta 2005, 26, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Louwen, F.; Muschol-Steinmetz, C.; Reinhard, J.; Reitter, A.; Yuan, J. A lesson for cancer research: Placental microarray gene analysis in preeclampsia. Oncotarget 2012, 3, 759–773. [Google Scholar] [CrossRef] [Green Version]
- Muschol-Steinmetz, C.; Friemel, A.; Kreis, N.N.; Reinhard, J.; Yuan, J.; Louwen, F. Function of survivin in trophoblastic cells of the placenta. PLoS ONE 2013, 8, e73337. [Google Scholar] [CrossRef] [Green Version]
- Graham, C.H.; Hawley, T.S.; Hawley, R.G.; MacDougall, J.R.; Kerbel, R.S.; Khoo, N.; Lala, P.K. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp. Cell Res. 1993, 206, 204–211. [Google Scholar] [CrossRef]
- Dash, P.R.; Cartwright, J.E.; Whitley, G.S. Nitric oxide inhibits polyamine-induced apoptosis in the human extravillous trophoblast cell line SGHPL-4. Hum. Reprod. 2003, 18, 959–968. [Google Scholar] [CrossRef] [Green Version]
- Kreis, N.N.; Friemel, A.; Zimmer, B.; Roth, S.; Rieger, M.A.; Rolle, U.; Louwen, F.; Yuan, J. Mitotic p21Cip1/CDKN1A is regulated by cyclin-dependent kinase 1 phosphorylation. Oncotarget 2016, 7, 50215–50228. [Google Scholar] [CrossRef] [Green Version]
- Kreis, N.N.; Sommer, K.; Sanhaji, M.; Kramer, A.; Matthess, Y.; Kaufmann, M.; Strebhardt, K.; Yuan, J. Long-term downregulation of Polo-like kinase 1 increases the cyclin-dependent kinase inhibitor p21(WAF1/CIP1). Cell Cycle 2009, 8, 460–472. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.Y.; Wang, L.J.; Chen, C.P.; Chen, L.F.; Chen, Y.H.; Chen, H. GCM1 regulation of the expression of syncytin 2 and its cognate receptor MFSD2A in human placenta. Biol. Reprod. 2010, 83, 387–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, A.B.; Tunster, S.J.; Savory, N.; Holmes, A.; Beasley, J.; Parveen, S.A.; Penketh, R.J.; John, R.M. Placental expression of imprinted genes varies with sampling site and mode of delivery. Placenta 2015, 36, 790–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, J.M.; Hubel, C.A. The two stage model of preeclampsia: Variations on the theme. Placenta 2009, 30, S32–S37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verlohren, S.; Geusens, N.; Morton, J.; Verhaegen, I.; Hering, L.; Herse, F.; Dudenhausen, J.W.; Muller, D.N.; Luft, F.C.; Cartwright, J.E.; et al. Inhibition of trophoblast-induced spiral artery remodeling reduces placental perfusion in rat pregnancy. Hypertension 2010, 56, 304–310. [Google Scholar] [CrossRef] [Green Version]
- Orendi, K.; Gauster, M.; Moser, G.; Meiri, H.; Huppertz, B. The choriocarcinoma cell line BeWo: Syncytial fusion and expression of syncytium-specific proteins. Reproduction 2010, 140, 759–766. [Google Scholar] [CrossRef] [Green Version]
- Coulombe, P.; Meloche, S. Atypical mitogen-activated protein kinases: Structure, regulation and functions. Biochim. Biophys. Acta 2007, 1773, 1376–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef] [Green Version]
- Sharp, A.N.; Heazell, A.E.; Baczyk, D.; Dunk, C.E.; Lacey, H.A.; Jones, C.J.; Perkins, J.E.; Kingdom, J.C.; Baker, P.N.; Crocker, I.P. Preeclampsia is associated with alterations in the p53-pathway in villous trophoblast. PLoS ONE 2014, 9, e87621. [Google Scholar] [CrossRef]
- Cobellis, L.; Mastrogiacomo, A.; Federico, E.; Schettino, M.T.; De Falco, M.; Manente, L.; Coppola, G.; Torella, M.; Colacurci, N.; De Luca, A. Distribution of Notch protein members in normal and preeclampsia-complicated placentas. Cell Tissue Res. 2007, 330, 527–534. [Google Scholar] [CrossRef]
- Louwen, F.; Muschol-Steinmetz, C.; Friemel, A.; Kampf, A.K.; Tottel, E.; Reinhard, J.; Yuan, J. Targeted gene analysis: Increased B-cell lymphoma 6 in preeclamptic placentas. Hum. Pathol. 2014, 45, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Londero, A.P.; Orsaria, M.; Marzinotto, S.; Grassi, T.; Fruscalzo, A.; Calcagno, A.; Bertozzi, S.; Nardini, N.; Stella, E.; Lelle, R.J.; et al. Placental aging and oxidation damage in a tissue micro-array model: An immunohistochemistry study. Histochem. Cell Biol. 2016, 146, 191–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, L.; Schimmelmann, M.; Wu, Y.; Reisch, B.; Faas, M.; Kimmig, R.; Winterhager, E.; Koninger, A.; Gellhaus, A. CCN3 Signaling Is Differently Regulated in Placental Diseases Preeclampsia and Abnormally Invasive Placenta. Front Endocrinol. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Genbacev, O.; Zhou, Y.; Ludlow, J.W.; Fisher, S.J. Regulation of human placental development by oxygen tension. Science 1997, 277, 1669–1672. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Longtine, M.S.; Sadovsky, Y.; Nelson, D.M. Hypoxia downregulates p53 but induces apoptosis and enhances expression of BAD in cultures of human syncytiotrophoblasts. Am. J. Physiol. Cell Physiol. 2010, 299, C968–C976. [Google Scholar] [CrossRef] [Green Version]
- Phan, R.T.; Saito, M.; Basso, K.; Niu, H.; Dalla-Favera, R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat. Immunol. 2005, 6, 1054–1060. [Google Scholar] [CrossRef]
- Sharma, N.; Kubaczka, C.; Kaiser, S.; Nettersheim, D.; Mughal, S.S.; Riesenberg, S.; Holzel, M.; Winterhager, E.; Schorle, H. Tpbpa-Cre-mediated deletion of TFAP2C leads to deregulation of Cdkn1a, Akt1 and the ERK pathway, causing placental growth arrest. Development 2016, 143, 787–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotani, T.; Iwase, A.; Ino, K.; Sumigama, S.; Yamamoto, E.; Hayakawa, H.; Nagasaka, T.; Itakura, A.; Nomura, S.; Kikkawa, F. Activator protein-2 impairs the invasion of a human extravillous trophoblast cell line. Endocrinology 2009, 150, 4376–4385. [Google Scholar] [CrossRef] [Green Version]
- Brock, M.; Haider, T.J.; Vogel, J.; Gassmann, M.; Speich, R.; Trenkmann, M.; Ulrich, S.; Kohler, M.; Huber, L.C. The hypoxia-induced microRNA-130a controls pulmonary smooth muscle cell proliferation by directly targeting CDKN1A. Int. J Biochem. Cell Biol. 2015, 61, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Li, H.; Ge, Q.; Guo, L.; Chen, F. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Mol. Med. Rep. 2015, 12, 527–534. [Google Scholar] [CrossRef] [Green Version]
- Kudo, Y.; Boyd, C.A.; Sargent, I.L.; Redman, C.W. Hypoxia alters expression and function of syncytin and its receptor during trophoblast cell fusion of human placental BeWo cells: Implications for impaired trophoblast syncytialisation in pre-eclampsia. Biochim. Biophys. Acta 2003, 1638, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Racca, A.C.; Ridano, M.E.; Camolotto, S.; Genti-Raimondi, S.; Panzetta-Dutari, G.M. A novel regulator of human villous trophoblast fusion: The Kruppel-like factor 6. Mol. Hum. Reprod. 2015, 21, 347–358. [Google Scholar] [CrossRef] [Green Version]
- Ullah, R.; Naz, A.; Akram, H.S.; Ullah, Z.; Tariq, M.; Mithani, A.; Faisal, A. Transcriptomic analysis reveals differential gene expression, alternative splicing, and novel exons during mouse trophoblast stem cell differentiation. Stem. Cell Res. Ther. 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gauster, M.; Maninger, S.; Siwetz, M.; Deutsch, A.; El-Heliebi, A.; Kolb-Lenz, D.; Hiden, U.; Desoye, G.; Herse, F.; Prokesch, A. Downregulation of p53 drives autophagy during human trophoblast differentiation. Cell Mol. Life Sci. 2018, 75, 1839–1855. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Wang, R.; Zhu, C.; Wang, H.; Lin, H.Y.; Gu, Y.; Cross, J.C.; Wang, H. Fine-Tuned and Cell-Cycle-Restricted Expression of Fusogenic Protein Syncytin-2 Maintains Functional Placental Syncytia. Cell Rep. 2017, 21, 1150–1159. [Google Scholar] [CrossRef] [Green Version]
- Vargas, A.; Toufaily, C.; LeBellego, F.; Rassart, E.; Lafond, J.; Barbeau, B. Reduced expression of both syncytin 1 and syncytin 2 correlates with severity of preeclampsia. Reprod. Sci. 2011, 18, 1085–1091. [Google Scholar] [CrossRef] [PubMed]
- Unek, G.; Ozmen, A.; Mendilcioglu, I.; Simsek, M.; Korgun, E.T. The expression of cell cycle related proteins PCNA, Ki67, p27 and p57 in normal and preeclamptic human placentas. Tissue Cell 2014, 46, 198–205. [Google Scholar] [CrossRef]
- Song, H.L.; Liu, T.H.; Wang, Y.H.; Li, F.F.; Ruan, L.L.; Adu-Gyamfi, E.A.; Hu, S.C.; Chen, X.M.; Ding, Y.B.; Fu, L.J. Appropriate expression of P57kip2 drives trophoblast fusion via cell cycle arrest. Reproduction 2021, 161, 633–644. [Google Scholar] [CrossRef]
- Takahashi, K.; Yoneyama, Y.; Koizumi, N.; Utoguchi, N.; Kanayama, N.; Higashi, N. Expression of p57(KIP2) reduces growth and invasion, and induces syncytialization in a human placental choriocarcinoma cell line, BeWo. Placenta 2021, 104, 168–178. [Google Scholar] [CrossRef] [PubMed]
- He, G.Q.; Liu, G.Y.; Xu, W.M.; Liao, H.J.; Liu, X.H.; He, G.L. p57KIP2mediated inhibition of human trophoblast apoptosis and promotion of invasion in vitro. Int. J. Mol. Med. 2019, 44, 281–290. [Google Scholar] [CrossRef]
- Kanayama, N.; Takahashi, K.; Matsuura, T.; Sugimura, M.; Kobayashi, T.; Moniwa, N.; Tomita, M.; Nakayama, K. Deficiency in p57Kip2 expression induces preeclampsia-like symptoms in mice. Mol. Hum. Reprod. 2002, 8, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Falcao, S.; Solomon, C.; Monat, C.; Berube, J.; Gutkowska, J.; Lavoie, J.L. Impact of diet and stress on the development of preeclampsia-like symptoms in p57kip2 mice. Am. J. Physiol. Heart Circ. Physiol. 2009, 296, H119–H126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knox, K.S.; Baker, J.C. Genome-wide expression profiling of placentas in the p57Kip2 model of pre-eclampsia. Mol. Hum. Reprod. 2007, 13, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Ullah, Z.; Kohn, M.J.; Yagi, R.; Vassilev, L.T.; DePamphilis, M.L. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev. 2008, 22, 3024–3036. [Google Scholar] [CrossRef] [Green Version]
- Engeland, K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018, 25, 114–132. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Quaas, M.; Steiner, L.; Engeland, K. The p53-p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016, 44, 164–174. [Google Scholar] [CrossRef] [Green Version]
- Rothbauer, M.; Patel, N.; Gondola, H.; Siwetz, M.; Huppertz, B.; Ertl, P. A comparative study of five physiological key parameters between four different human trophoblast-derived cell lines. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Yoshihara, E.; Masaki, S.; Matsuo, Y.; Chen, Z.; Tian, H.; Yodoi, J. Thioredoxin/Txnip: Redoxisome, as a redox switch for the pathogenesis of diseases. Front Immunol. 2014, 4, 514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mogami, H.; Yura, S.; Kondoh, E.; Masutani, H.; Yodoi, J.; Konishi, I. Differential expression of thioredoxin binding protein-2/Txnip in human placenta: Possible involvement of hypoxia in its suppression during early pregnancy. J. Obstet. Gynaecol. Res. 2017, 43, 50–56. [Google Scholar] [CrossRef] [Green Version]
- Ji Cho, M.; Yoon, S.J.; Kim, W.; Park, J.; Lee, J.; Park, J.G.; Cho, Y.L.; Hun Kim, J.; Jang, H.; Park, Y.J.; et al. Oxidative stress-mediated TXNIP loss causes RPE dysfunction. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, J.H.; Lee, K.N.; Hwang, C.Y.; Kwon, K.S.; You, K.H.; Choi, I. Tumor suppressor VDUP1 increases p27(kip1) stability by inhibiting JAB1. Cancer Res. 2005, 65, 4485–4489. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| n | Gestational Age (Weeks) | Body Mass Index (BMI) | Age | Birth Weight (g) | |
|---|---|---|---|---|---|
| CS | 5 | 38.2 ± 0.8 | 21.9 ± 1.4 | 28.4 ± 0.9 | 2960 ± 634 |
| eCS | 5 | 38.0 ± 1.0 | 21.5 ± 1.9 | 27.0 ± 2.2 | 3020 ± 370 |
| VD | 5 | 38.0 ± 1.0 | 21.7 ± 2.3 | 27.0 ± 1.9 | 3002 ± 302 |
| opVD | 5 | 38.0 ± 1.0 | 22.8 ± 1.0 | 26.4 ± 2.2 | 3008 ± 302 |
| p-value | ns | ns | ns | ns |
| n | Age | Gestational Age (Weeks) | Body Mass Index (BMI) | Birth Weight (g) | Systolic BP (mmHg) | Diastolic BP (mmHg) | Proteinuria (mg/24 h) | |
|---|---|---|---|---|---|---|---|---|
| control | 20 | 32.6 ± 4.6 | 29.7 ± 2.6 | 24.9 ± 3.9 | 1284 ± 710 | 118 ± 13 | 71 ± 11 | n.d. |
| early-onset PE | 20 | 32.4 ± 5.9 | 29.6 ± 2.6 | 25.7 ± 4.4 | 1072 ± 387 | 167 ± 22 | 102 ± 12 | 4153 ± 4569 |
| p-value | 0.876 | 0.330 | 0.256 | 0.114 | 0.00000011 | 0.0000038 |
| n | Age | Gestational Age (Weeks) | Body Mass Index (BMI) | Birth Weight (g) | Systolic BP (mmHg) | Diastolic BP (mmHg) | Proteinuria (mg/24 h) | |
|---|---|---|---|---|---|---|---|---|
| control | 10 | 30.9 ± 3.4 | 37.7 ± 1.3 | 23.4 ± 4.0 | 2914 ± 524 | 119 ± 7 | 73 ± 11 | n.d. |
| late-onset PE | 10 | 30.9 ± 2.8 | 37.7 ± 1.3 | 24.4 ± 2.0 | 2413 ± 461 | 153 ± 16 | 96 ± 13 | 1794 ± 1901 |
| p-value | 0.989 | 1.0 | 0.273 | 0.020 | 0.00034 | 0.00020 |
| n | Age | Gestational Age (Weeks) | Body Mass Index (BMI) | Birth Weight (g) | Systolic BP (mmHg) | Diastolic BP (mmHg) | Proteinuria (mg/24 h) | |
|---|---|---|---|---|---|---|---|---|
| control | 17 | 29.6 ± 9.1 | 30.2 ± 7.9 | 23.7 ± 6.7 | 1751 ± 920 | 109 ± 29 | 66 ± 20 | n.d. |
| ePE + HELLP | 16 | 31.4 ± 4.5 | 31.7 ± 2.4 | 25.3 ± 4.3 | 1389 ± 443 | 175 ± 21 | 107 ± 9 | 4815 ± 4856 |
| p-value | 0.589 | 0.164 | 0.973 | 0.0035 | 0.0000001 | 0.00000004 |
| n | Age | Gestational Age (Weeks) | Body Mass Index (BMI) | Birth Weight (g) | Systolic BP (mmHg) | Diastolic BP (mmHg) | Proteinuria (mg/24 h) | |
|---|---|---|---|---|---|---|---|---|
| control | 5 | 32.6 ± 5.3 | 40 ± 1.2 | 22.6 ± 3.7 | 3407 ± 448 | 114 ± 9 | 74 ± 11 | n.d. |
| PE | 5 | 34.1 ± 5 | 36.8 ± 4.0 | 23.7 ± 2.7 | 2462 ± 974 | 150 ± 11 | 97 ± 5 | 2763 ± 3459 |
| p-value | 0.666 | 0.123 | 0.604 | 0.084 | 0.00042 | 0.0031 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kreis, N.-N.; Friemel, A.; Jennewein, L.; Hoock, S.C.; Hentrich, A.E.; Nowak, T.; Louwen, F.; Yuan, J. Functional Analysis of p21Cip1/CDKN1A and Its Family Members in Trophoblastic Cells of the Placenta and Its Roles in Preeclampsia. Cells 2021, 10, 2214. https://doi.org/10.3390/cells10092214
Kreis N-N, Friemel A, Jennewein L, Hoock SC, Hentrich AE, Nowak T, Louwen F, Yuan J. Functional Analysis of p21Cip1/CDKN1A and Its Family Members in Trophoblastic Cells of the Placenta and Its Roles in Preeclampsia. Cells. 2021; 10(9):2214. https://doi.org/10.3390/cells10092214
Chicago/Turabian StyleKreis, Nina-Naomi, Alexandra Friemel, Lukas Jennewein, Samira Catharina Hoock, Anna Elisabeth Hentrich, Thorsten Nowak, Frank Louwen, and Juping Yuan. 2021. "Functional Analysis of p21Cip1/CDKN1A and Its Family Members in Trophoblastic Cells of the Placenta and Its Roles in Preeclampsia" Cells 10, no. 9: 2214. https://doi.org/10.3390/cells10092214
APA StyleKreis, N. -N., Friemel, A., Jennewein, L., Hoock, S. C., Hentrich, A. E., Nowak, T., Louwen, F., & Yuan, J. (2021). Functional Analysis of p21Cip1/CDKN1A and Its Family Members in Trophoblastic Cells of the Placenta and Its Roles in Preeclampsia. Cells, 10(9), 2214. https://doi.org/10.3390/cells10092214