Dopamine Reduces SARS-CoV-2 Replication In Vitro through Downregulation of D2 Receptors and Upregulation of Type-I Interferons
Abstract
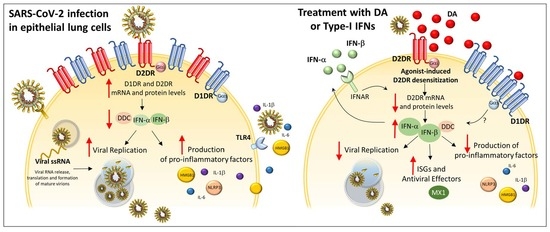
:1. Introduction
2. Materials and Methods
2.1. Cell Lines, Virus, and Reagents
2.2. In Vitro SARS-CoV-2 Infection Assay and Treatment Protocols
2.3. RNA Extraction, Reverse Transcription, and Gene Expression
2.3.1. Viral Replication
2.3.2. Cellular RNA Extraction, Reverse Transcription, and Gene Expression
2.4. Immunocytochemistry
2.5. MTT and Trypan Blue Cell Viability Assays
2.6. Statistics
3. Results
3.1. DA in the Micromolar Range Reduces SARS-CoV-2 Replication in CaLu-3 Cells
3.2. Dopamine Receptor Agonists Recapitulate While Dopamine Receptor Antagonists Reverse DA-Induced Inhibition of SARS-CoV-2 Replication
3.3. The Antiviral Effect of DA Is Associated with Downregulation of D2DRs
3.4. The Antiviral Effect of DA and D2DR Agonist Is Associated with Upregulation of Type-I Interferons and Downregulation of Pro-Inflammatory Mediators
3.5. Type-I Interferons Suppress SARS-CoV-2 Replication, Which Is Associated with Downregulation of D2DRs and the Rescue of DA Synthetic Enzymes
3.6. A549-hACE2 Lack D2DRs and Are Less Susceptible to Both SARS-CoV-2 Infection and Type-IFN Treatment
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CREB | cAMP Response Element-Binding protein |
| D1DR | D1 Dopamine Receptor |
| D2DR | D2 Dopamine Receptor |
| DA | Dopamine |
| DAT | Dopamine Transporter |
| DDC | L-Dopa-Decarboxylase |
| GSK3-β | Glycogen Synthase Kinase 3-beta |
| HMGB1 | High-Mobility Group Box 1 |
| IFN | Interferon |
| IL-1b | Interleukin 1 beta |
| IL-6 | Interleukin 6 |
| IRF3 | Interferon Regulatory Factor 3 |
| MAPK3/ERK1 | Mitogen-activated Protein Kinase 3 |
| MOI | Multiplicity of Infection |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| MX-A | Human myxovirus resistance protein 1 |
| NLRP3 | NLR family pyrin domain containing 3 Inflammasome |
| PKA | cAMP-dependent Protein Kinase A |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome-Coronavirus-2 |
| TCID50 | 50% Tissue Culture Infectious Dose |
| TH | Tyrosine Hydroxylase |
| TLR4 | Toll-like Receptor 4 |
References
- Khalefah, M.M.; Khalifah, A.M. Determining the relationship between SARS-CoV-2 infection, dopamine, and COVID-19 complications. J. Taibah Univ. Med. Sci. 2020, 15, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Nataf, S. An alteration of the dopamine synthetic pathway is possibly involved in the pathophysiology of COVID-19. J. Med. Virol. 2020, 92, 1743–1744. [Google Scholar] [CrossRef] [PubMed]
- Nataf, S.; Pays, L. Molecular Insights into SARS-CoV2-Induced Alterations of the Gut/Brain Axis. Int. J. Mol. Sci. 2021, 22, 10440. [Google Scholar] [CrossRef] [PubMed]
- Brundin, P.; Nath, A.; Beckham, J.D. Is COVID-19 a Perfect Storm for Parkinson’s Disease? Trends Neurosci. 2020, 43, 931–933. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Jiang, W.; Liu, L.; Wang, X.; Ding, C.; Tian, Z.; Zhou, R. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 2015, 160, 62–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matt, S.M.; Gaskill, P.J. Where Is Dopamine and how do Immune Cells See it?: Dopamine-Mediated Immune Cell Function in Health and Disease. J. Neuroimmune Pharmacol. 2020, 15, 114–164. [Google Scholar] [CrossRef]
- Limanaqi, F.; Busceti, C.L.; Biagioni, F.; Lazzeri, G.; Forte, M.; Schiavon, S.; Sciarretta, S.; Frati, G.; Fornai, F. Cell Clearing Systems as Targets of Polyphenols in Viral Infections: Potential Implications for COVID-19 Pathogenesis. Antioxidants 2020, 9, 1105. [Google Scholar] [CrossRef]
- Sarkar, C.; Basu, B.; Chakroborty, D.; Dasgupta, P.S.; Basu, S. The immunoregulatory role of dopamine: An update. Brain Behav. Immun. 2010, 24, 525–528. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Lu, Y. Immunomodulatory Effects of Dopamine in Inflammatory Diseases. Front. Immunol. 2021, 12, 663102. [Google Scholar] [CrossRef]
- Limanaqi, F.; Biagioni, F.; Busceti, C.L.; Ryskalin, L.; Soldani, P.; Frati, A.; Fornai, F. Cell Clearing Systems Bridging Neuro-Immunity and Synaptic Plasticity. Int. J. Mol. Sci. 2019, 20, 2197. [Google Scholar] [CrossRef] [Green Version]
- Martel, J.C.; Gatti McArthur, S. Dopamine Receptor Subtypes, Physiology and Pharmacology: New Ligands and Concepts in Schizophrenia. Front. Pharmacol. 2020, 11, 1003. [Google Scholar] [CrossRef] [PubMed]
- Bertorello, A.M.; Sznajder, J.I. The dopamine paradox in lung and kidney epithelia: Sharing the same target but operating different signaling networks. Am. J. Respir. Cell Mol. Biol. 2005, 33, 432–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chua, B.A.; Perks, A.M. The effect of dopamine on lung liquid production by in vitro lungs from fetal guinea-pigs. J. Physiol. 1998, 513, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Baig, A.M.; Khaleeq, A.; Syeda, H.; Bibi, N. Docking Prediction of Levodopa in the Receptor Binding Domain of Spike Protein of SARS-CoV-2. ACS Pharmacol. Transl. Sci. 2021, 4, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Bibi, N.; Farid, A.; Gul, S.; Ali, J.; Amin, F.; Kalathiya, U.; Hupp, T. Drug repositioning against COVID-19: A first line treatment. J. Biomol. Struct. Dyn. 2021, 1–15. [Google Scholar] [CrossRef]
- Chan, S.W.; Shafi, T.; Ford, R.C. Kite-Shaped Molecules Block SARS-CoV-2 Cell Entry at a Post-Attachment Step. Viruses 2021, 13, 2306. [Google Scholar] [CrossRef]
- Fink, K.; Nitsche, A.; Neumann, M.; Grossegesse, M.; Eisele, K.H.; Danysz, W. Amantadine Inhibits SARS-CoV-2 In Vitro. Viruses 2021, 13, 539. [Google Scholar] [CrossRef]
- Rejdak, K.; Grieb, P. Adamantanes might be protective from COVID-19 in patients with neurological diseases: Multiple sclerosis, parkinsonism and cognitive impairment. Mult. Scler. Relat. Disord. 2020, 42, 102163. [Google Scholar] [CrossRef]
- Barrow, K.A.; Rich, L.M.; Vanderwall, E.R.; Reeves, S.R.; Rathe, J.A.; White, M.P.; Debley, J.S. Inactivation of Material from SARS-CoV-2-Infected Primary Airway Epithelial Cell Cultures. Methods Protoc. 2021, 4, 7. [Google Scholar] [CrossRef]
- Saulle, I.; Vanetti, C.; Goglia, S.; Vicentini, C.; Tombetti, E.; Garziano, M.; Clerici, M.; Biasin, M. A New ERAP2/Iso3 Isoform Expression Is Triggered by Different Microbial Stimuli in Human Cells. Could It Play a Role in the Modulation of SARS-CoV-2 Infection? Cells 2020, 9, 1951. [Google Scholar] [CrossRef]
- Trabattoni, D.; Gnudi, F.; Ibba, S.V.; Saulle, I.; Agostini, S.; Masetti, M.; Biasin, M.; Rossignol, J.F.; Clerici, M. Thiazolides Elicit Anti-Viral Innate Immunity and Reduce HIV Replication. Sci. Rep. 2016, 6, 27148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirigiti, P.; Bai, Y.; Yang, Y.F.; Li, X.; Li, B.; Brewer, G.; Machida, C.A. Agonist-mediated down-regulation of rat beta1-adrenergic receptor transcripts: Role of potential post-transcriptional degradation factors. Mol. Pharmacol. 2001, 60, 1308–1324. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Aloyo, V.J.; Weiss, B. Continuous treatment with the D2 dopamine receptor agonist quinpirole decreases D2 dopamine receptors, D2 dopamine receptor messenger RNA and proenkephalin messenger RNA, and increases mu opioid receptors in mouse striatum. Neuroscience 1993, 54, 669–680. [Google Scholar] [CrossRef]
- Tsai, S.J. Dopamine receptor downregulation: An alternative strategy for schizophrenia treatment. Med. Hypotheses 2004, 63, 1047–1050. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ji, X.; Liu, J.; Li, Z.; Zhang, X. Dopamine Receptor Subtypes Differentially Regulate Autophagy. Int. J. Mol. Sci. 2018, 19, 1540. [Google Scholar] [CrossRef] [Green Version]
- Haak, A.J.; Kostallari, E.; Sicard, D.; Ligresti, G.; Choi, K.M.; Caporarello, N.; Jones, D.L.; Tan, Q.; Meridew, J.; Diaz Espinosa, A.M.; et al. Selective YAP/TAZ inhibition in fibroblasts via dopamine receptor D1 agonism reverses fibrosis. Sci. Transl. Med. 2019, 11, eaau6296. [Google Scholar] [CrossRef]
- Ciarka, A.; Vincent, J.L.; van de Borne, P. The effects of dopamine on the respiratory system: Friend or foe? Pulm. Pharmacol. Ther. 2007, 20, 607–615. [Google Scholar] [CrossRef] [Green Version]
- Gaskill, P.J.; Yano, H.H.; Kalpana, G.V.; Javitch, J.A.; Berman, J.W. Dopamine receptor activation increases HIV entry into primary human macrophages. PLoS ONE 2014, 9, e108232. [Google Scholar] [CrossRef] [Green Version]
- Simanjuntak, Y.; Liang, J.J.; Lee, Y.L.; Lin, Y.L. Japanese Encephalitis Virus Exploits Dopamine D2 Receptor-phospholipase C to Target Dopaminergic Human Neuronal Cells. Front. Microbiol. 2017, 8, 651. [Google Scholar] [CrossRef]
- Ferone, D.; Arvigo, M.; Semino, C.; Jaquet, P.; Saveanu, A.; Taylor, J.E.; Moreau, J.P.; Culler, M.D.; Albertelli, M.; Minuto, F.; et al. Somatostatin and dopamine receptor expression in lung carcinoma cells and effects of chimeric somatostatin-dopamine molecules on cell proliferation. Am. J. Physiol. Endocrinol. Metab. 2005, 289, E1044–E1050. [Google Scholar] [CrossRef]
- Klijn, C.; Durinck, S.; Stawiski, E.W.; Haverty, P.M.; Jiang, Z.; Liu, H.; Degenhardt, J.; Mayba, O.; Gnad, F.; Liu, J.; et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015, 33, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Helms, M.N.; Chen, X.J.; Ramosevac, S.; Eaton, D.C.; Jain, L. Dopamine regulation of amiloride-sensitive sodium channels in lung cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2006, 290, L710–L722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campa, D.; Zienolddiny, S.; Lind, H.; Ryberg, D.; Skaug, V.; Canzian, F.; Haugen, A. Polymorphisms of dopamine receptor/transporter genes and risk of non-small cell lung cancer. Lung Cancer 2007, 56, 17–23. [Google Scholar] [CrossRef]
- Miyajima, K.; Kawamoto, C.; Hara, S.; Mori-Kojima, M.; Ohye, T.; Sumi-Ichinose, C.; Saito, N.; Sasaoka, T.; Metzger, D.; Ichinose, H. Tyrosine hydroxylase conditional KO mice reveal peripheral tissue-dependent differences in dopamine biosynthetic pathways. J. Biol. Chem. 2021, 296, 100544. [Google Scholar] [CrossRef] [PubMed]
- Mukaetova-Ladinska, E.B.; Kronenberg, G.; Raha-Chowdhury, R. COVID-19 and neurocognitive disorders. Curr. Opin. Psychiatry 2021, 34, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Owesson-White, C.A.; Roitman, M.F.; Sombers, L.A.; Belle, A.M.; Keithley, R.B.; Peele, J.L.; Carelli, R.M.; Wightman, R.M. Sources contributing to the average extracellular concentration of dopamine in the nucleus accumbens. J. Neurochem. 2012, 121, 252–262. [Google Scholar] [CrossRef]
- Kawano, M.; Takagi, R.; Saika, K.; Matsui, M.; Matsushita, S. Dopamine regulates cytokine secretion during innate and adaptive immune responses. Int. Immunol. 2018, 30, 591–606. [Google Scholar] [CrossRef]
- Saldías, F.J.; Comellas, A.P.; Pesce, L.; Lecuona, E.; Sznajder, J.I. Dopamine increases lung liquid clearance during mechanical ventilation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002, 283, L136–L143. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Reith, M.E.A.; Liu-Chen, L.Y.; Kortagere, S. Biased signaling agonist of dopamine D3 receptor induces receptor internalization independent of β-arrestin recruitment. Pharmacol. Res. 2019, 143, 48–57. [Google Scholar] [CrossRef]
- Barton, A.C.; Black, L.E.; Sibley, D.R. Agonist-induced desensitization of D2 dopamine receptors in human Y-79 retinoblastoma cells. Mol. Pharmacol. 1991, 39, 650–658. [Google Scholar] [CrossRef]
- Bartlett, S.E.; Enquist, J.; Hopf, F.W.; Lee, J.H.; Gladher, F.; Kharazia, V.; Waldhoer, M.; Mailliard, W.S.; Armstrong, R.; Bonci, A.; et al. Dopamine responsiveness is regulated by targeted sorting of D2 receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 11521–11526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skinbjerg, M.; Ariano, M.A.; Thorsell, A.; Heilig, M.; Halldin, C.; Innis, R.B.; Sibley, D.R. Arrestin3 mediates D(2) dopamine receptor internalization. Synapse 2009, 63, 621–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thibault, D.; Albert, P.R.; Pineyro, G.; Trudeau, L.É. Neurotensin triggers dopamine D2 receptor desensitization through a protein kinase C and beta-arrestin1-dependent mechanism. J. Biol. Chem. 2011, 286, 9174–9184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Namkung, Y.; Dipace, C.; Javitch, J.A.; Sibley, D.R. G protein-coupled receptor kinase-mediated phosphorylation regulates post-endocytic trafficking of the D2 dopamine receptor. J. Biol. Chem. 2009, 284, 15038–15051. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Verma, A.; Garcia, G., Jr.; Ramage, H.; Lucas, A.; Myers, R.L.; Michaelson, J.J.; Coryell, W.; Kumar, A.; Charney, A.W.; et al. Targeting the coronavirus nucleocapsid protein through GSK-3 inhibition. Proc. Natl. Acad. Sci. USA 2021, 118, e2113401118. [Google Scholar] [CrossRef]
- Dunigan, C.D.; Hoang, Q.; Curran, P.K.; Fishman, P.H. Complexity of agonist- and cyclic AMP-mediated downregulation of the human beta 1-adrenergic receptor: Role of internalization, degradation, and mRNA destabilization. Biochemistry 2002, 41, 8019–8030. [Google Scholar] [CrossRef]
- Kotowski, S.J.; Hopf, F.W.; Seif, T.; Bonci, A.; von Zastrow, M. Endocytosis promotes rapid dopaminergic signaling. Neuron 2011, 71, 278–290. [Google Scholar] [CrossRef] [Green Version]
- Ford, C.P. The role of D2-autoreceptors in regulating dopamine neuron activity and transmission. Neuroscience 2014, 282, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Kuang, M.; Li, J.; Zhu, L.; Jia, Z.; Guo, X.; Hu, Y.; Kong, J.; Yin, H.; Wang, X.; et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021, 31, 818–820. [Google Scholar] [CrossRef]
- Aboudounya, M.M.; Heads, R.J. COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Mediators Inflamm. 2021, 2021, 8874339. [Google Scholar] [CrossRef]
- Sohn, K.M.; Lee, S.G.; Kim, H.J.; Cheon, S.; Jeong, H.; Lee, J.; Kim, I.S.; Silwal, P.; Kim, Y.J.; Paik, S.; et al. COVID-19 Patients Upregulate Toll-like Receptor 4-mediated Inflammatory Signaling That Mimics Bacterial Sepsis. J. Korean Med. Sci. 2020, 35, e343. [Google Scholar] [CrossRef] [PubMed]
- Brandão, S.C.S.; Ramos, J.O.X.; Dompieri, L.T.; Godoi, E.T.A.M.; Figueiredo, J.L.; Sarinho, E.S.C.; Chelvanambi, S.; Aikawa, M. Is Toll-like receptor 4 involved in the severity of COVID-19 pathology in patients with cardiometabolic comorbidities? Cytokine Growth Factor Rev. 2021, 58, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Andersson, U.; Ottestad, W.; Tracey, K.J. Extracellular HMGB1: A therapeutic target in severe pulmonary inflammation including COVID-19? Mol. Med. 2020, 26, 42. [Google Scholar] [CrossRef] [PubMed]
- Bombaci, G.; Sarangdhar, M.A.; Andina, N.; Tardivel, A.; Yu, E.C.; Mackie, G.M.; Pugh, M.; Ozan, V.B.; Banz, Y.; Spinetti, T.; et al. LRR-protein RNH1 dampens the inflammasome activation and is associated with COVID-19 severity. Life Sci. Alliance 2022, 5, e202101226. [Google Scholar] [CrossRef]
- Zhu, J.; Hu, Z.; Han, X.; Wang, D.; Jiang, Q.; Ding, J.; Xiao, M.; Wang, C.; Lu, M.; Hu, G. Dopamine D2 receptor restricts astrocytic NLRP3 inflammasome activation via enhancing the interaction of β-arrestin2 and NLRP3. Cell Death Differ. 2018, 25, 2037–2049. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Felgenhauer, U.; Schoen, A.; Gad, H.H.; Hartmann, R.; Schaubmar, A.R.; Failing, K.; Drosten, C.; Weber, F. Inhibition of SARS-CoV-2 by type I and type III interferons. J. Biol. Chem. 2020, 295, 13958–13964. [Google Scholar] [CrossRef]
- Mantlo, E.; Bukreyeva, N.; Maruyama, J.; Paessler, S.; Huang, C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020, 179, 104811. [Google Scholar] [CrossRef]
- Busnadiego, I.; Fernbach, S.; Pohl, M.O.; Karakus, U.; Huber, M.; Trkola, A.; Stertz, S.; Hale, B.G. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. mBio 2020, 11, e01928-20. [Google Scholar] [CrossRef]
- Lokugamage, K.G.; Hage, A.; de Vries, M.; Valero-Jimenez, A.M.; Schindewolf, C.; Dittmann, M.; Rajsbaum, R.; Menachery, V.D. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020, 94, e01410-20. [Google Scholar] [CrossRef]
- Ejlerskov, P.; Hultberg, J.G.; Wang, J.; Carlsson, R.; Ambjørn, M.; Kuss, M.; Liu, Y.; Porcu, G.; Kolkova, K.; Friis Rundsten, C.; et al. Lack of Neuronal IFN-β-IFNAR Causes Lewy Body- and Parkinson’s Disease-like Dementia. Cell 2015, 163, 324–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayn, M.; Hirschenberger, M.; Koepke, L.; Nchioua, R.; Straub, J.H.; Klute, S.; Hunszinger, V.; Zech, F.; Prelli Bozzo, C.; Aftab, W.; et al. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 2021, 35, 109126. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.Q.; Zhuang, L.J.; Bao, H.Q.; Li, S.J.; Dai, F.Y.; Wang, P.; Li, Q.; Yin, D.M. Olfactory regulation by dopamine and DRD2 receptor in the nose. Proc. Natl. Acad. Sci. USA 2022, 119, e2118570119. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Qin, D.; Lv, H.; Fan, W.; Zhou, F.; Gao, Z.; Tao, Z.; Xu, Y. Activation of Dopamine D2 Receptor Alleviates Neuroinflammation in a Mouse Model of Allergic Rhinitis with Olfactory Dysfunction. Allergy Asthma Immunol. Res. 2021, 13, 882–895. [Google Scholar] [CrossRef]
- Mishra, A.; Singh, S.; Shukla, S. Physiological and Functional Basis of Dopamine Receptors and Their Role in Neurogenesis: Possible Implication for Parkinson’s disease. J. Exp. Neurosci. 2018, 12, 1179069518779829. [Google Scholar] [CrossRef] [Green Version]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Limanaqi, F.; Zecchini, S.; Dino, B.; Strizzi, S.; Cappelletti, G.; Utyro, O.; Vanetti, C.; Garziano, M.; Saulle, I.; Clerici, M.; et al. Dopamine Reduces SARS-CoV-2 Replication In Vitro through Downregulation of D2 Receptors and Upregulation of Type-I Interferons. Cells 2022, 11, 1691. https://doi.org/10.3390/cells11101691
Limanaqi F, Zecchini S, Dino B, Strizzi S, Cappelletti G, Utyro O, Vanetti C, Garziano M, Saulle I, Clerici M, et al. Dopamine Reduces SARS-CoV-2 Replication In Vitro through Downregulation of D2 Receptors and Upregulation of Type-I Interferons. Cells. 2022; 11(10):1691. https://doi.org/10.3390/cells11101691
Chicago/Turabian StyleLimanaqi, Fiona, Silvia Zecchini, Borana Dino, Sergio Strizzi, Gioia Cappelletti, Olga Utyro, Claudia Vanetti, Micaela Garziano, Irma Saulle, Mario Clerici, and et al. 2022. "Dopamine Reduces SARS-CoV-2 Replication In Vitro through Downregulation of D2 Receptors and Upregulation of Type-I Interferons" Cells 11, no. 10: 1691. https://doi.org/10.3390/cells11101691
APA StyleLimanaqi, F., Zecchini, S., Dino, B., Strizzi, S., Cappelletti, G., Utyro, O., Vanetti, C., Garziano, M., Saulle, I., Clerici, M., & Biasin, M. (2022). Dopamine Reduces SARS-CoV-2 Replication In Vitro through Downregulation of D2 Receptors and Upregulation of Type-I Interferons. Cells, 11(10), 1691. https://doi.org/10.3390/cells11101691