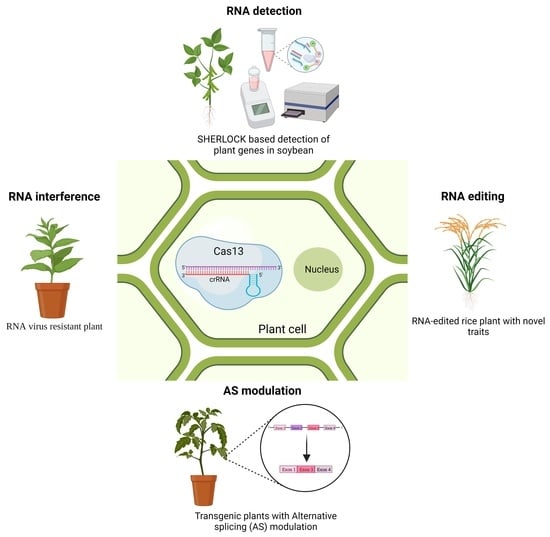
Applications of CRISPR/Cas13-Based RNA Editing in Plants
Abstract
:1. CRISPR/Cas Systems
2. Overview of CRISPR/Cas13 Systems
CRISPR/Cas13 Classification
| Type of Cas13 | Orthologues | Structural Composition | Functional Region | Application Scope | Reference |
|---|---|---|---|---|---|
| Cas13a (1250 aa) | LshCas13a | HEPN Domains (center and C terminus), pre-CrRNA processing, 3′ Non-G PFS preference (except Lwa and LbuCas13a), DR present on the 5′ end | ssRNA (spacer length 28–30 nt) | Virus resistance, RNA knockdown, disease diagnostics | [21,24,26,32] |
| LseCas13a | |||||
| LwaCas13a | |||||
| LbuCas13a | |||||
| LbaCas13a | |||||
| Cas13b (1150 aa) | BzCas13b | HEPN Domains (N and C terminus), pre-CrRNA processing, 5′ PFS preference of D, 3′ PFS NAN/NNA (Except PspCas13b), DR present on 3′ end | ssRNA (spacer length 30 nt) | Virus resistance, RNA base editing, RNA knockdown | [17,33] |
| PguCas13b | |||||
| PspCas13b | |||||
| Cas13c (1120 aa) | FpeCas13c | HEPN domains (center and C terminus), pre-CrRNA processing, No PFS preference, DR present on 5′ end | ssRNA (spacer length 28–30 nt) | RNA knockdown | [19,28] |
| Cas13d (930 aa) | RfxCas13d | HEPN domains (center and C terminus), pre-CrRNA processing, No PFS preference, DR present on 5′ end | ssRNA (spacer length 23–30 nt) | Virus resistance, RNA knockdown, alternative splicing modulation | [34,35] |
3. Applications of CRISPR/Cas13 Systems in Plants
3.1. RNA Interference against Viruses
3.2. RNA Targeting/Knockdown
3.3. RNA Editing
3.4. Modulation of Alternative Splicing
3.5. RNA Tracking and Nucleic Acid Detection
4. Prospective Directions of CRISPR/Cas13 RNA Targeting Systems in Plants
5. Potential Limitations of CRISPR/Cas13
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jansen, R.; Embden, J.D.V.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and Archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Classification and nomenclature of CRISPR-Cas systems: Where from here? CRISPR J. 2018, 1, 325–336. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Li, D.; Qiu, Z.; Shao, Y.; Chen, Y.; Guan, Y.; Liu, M.; Li, Y.; Gao, N.; Wang, L.; Lu, X. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 681–683. [Google Scholar] [CrossRef]
- Xie, K.; Yang, Y. RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef]
- Zhang, H.; Cheng, Q.-X.; Liu, A.-M.; Zhao, G.-P.; Wang, J. A novel and efficient method for bacteria genome editing employing both CRISPR/Cas9 and an antibiotic resistance cassette. Front. Microbiol. 2017, 8, 812. [Google Scholar] [CrossRef]
- Sretenovic, S.; Qi, Y. Plant prime editing goes prime. Nat. Plants 2022, 8, 20–22. [Google Scholar] [CrossRef]
- Zhou, C.; Zhou, H.; Ma, X.; Yang, H.; Wang, P.; Wang, G.; Zheng, L.; Zhang, Y.; Liu, X. Genome-wide identification and characterization of main histone modifications in Sorghum decipher regulatory mechanisms involved by mRNA and long noncoding RNA genes. J. Agric. Food Chem. 2021, 69, 2337–2347. [Google Scholar] [CrossRef] [PubMed]
- Uddin, F.; Rudin, C.M.; Sen, T. CRISPR gene therapy: Applications, limitations, and implications for the future. Front. Oncol. 2020, 10, 1387. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Wei, X.; Sheng, Z.; Hu, P.; Tang, S. CRISPR/Cas9 for development of disease resistance in plants: Recent progress, limitations and future prospects. Brief. Funct. Genom. 2020, 19, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Wolter, F.; Puchta, H. The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J. 2018, 94, 767–775. [Google Scholar] [CrossRef]
- Sharma, V.K.; Marla, S.; Zheng, W.; Mishra, D.; Huang, J.; Zhang, W.; Morris, G.P.; Cook, D.E. CRISPR guides induce gene silencing in plants in the absence of Cas. Genome Biol. 2022, 23, 6. [Google Scholar] [CrossRef]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef]
- Smargon, A.A.; Cox, D.B.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630. [Google Scholar] [CrossRef]
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 2018, 70, 327–339. [Google Scholar] [CrossRef]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef]
- Xu, C.; Zhou, Y.; Xiao, Q.; He, B.; Geng, G.; Wang, Z.; Cao, B.; Dong, X.; Bai, W.; Wang, Y. Programmable RNA editing with compact CRISPR–Cas13 systems from uncultivated microbes. Nat. Methods 2021, 18, 499–506. [Google Scholar] [CrossRef]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 2018, 173, 665–676. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.R. Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J. Mol. Biol. 2019, 431, 66–87. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.; Kellner, M.J.; Regev, A. RNA targeting with CRISPR–Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef]
- Knott, G.J.; East-Seletsky, A.; Cofsky, J.C.; Holton, J.M.; Charles, E.; O’Connell, M.R.; Doudna, J.A. Guide-bound structures of an RNA-targeting A-cleaving CRISPR–Cas13a enzyme. Nat. Struct. Mol. Biol. 2017, 24, 825–833. [Google Scholar] [CrossRef]
- Cox, D.B.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Huynh, N.; Depner, N.; Larson, R.; King-Jones, K. A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol. 2020, 21, 279. [Google Scholar] [CrossRef]
- He, B.; Peng, W.; Huang, J.; Zhang, H.; Zhou, Y.; Yang, X.; Liu, J.; Li, Z.; Xu, C.; Xue, M. Modulation of metabolic functions through Cas13d-mediated gene knockdown in liver. Protein Cell 2020, 11, 518. [Google Scholar] [CrossRef]
- Mahas, A.; Aman, R.; Mahfouz, M. CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol. 2019, 20, 263. [Google Scholar] [CrossRef] [Green Version]
- Bi, D.; Yao, J.; Wang, Y.; Qin, G.; Zhang, Y.; Wang, Y.; Zhao, J. CRISPR/Cas13d mediated efficient KDM5B mRNA knockdown in porcine cells and parthenogenetic embryos. Reproduction 2021, 162, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Kellner, M.J.; Zhang, F. Nucleic acid detection of plant genes using CRISPR-Cas13. CRISPR J. 2019, 2, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell 2019, 76, 826–837. [Google Scholar] [CrossRef]
- Wessels, H.-H.; Méndez-Mancilla, A.; Guo, X.; Legut, M.; Daniloski, Z.; Sanjana, N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 2020, 38, 722–727. [Google Scholar] [CrossRef]
- Du, M.; Jillette, N.; Zhu, J.J.; Li, S.; Cheng, A.W. CRISPR artificial splicing factors. Nat. Commun. 2020, 11, 2973. [Google Scholar] [CrossRef] [PubMed]
- Kordyś, M.; Sen, R.; Warkocki, Z. Applications of the versatile CRISPR-Cas13 RNA targeting system. Wiley Interdiscip. Rev. RNA 2021, 13, e1694. [Google Scholar] [CrossRef]
- Wang, F.; Wang, L.; Zou, X.; Duan, S.; Li, Z.; Deng, Z.; Luo, J.; Lee, S.Y.; Chen, S. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol. Adv. 2019, 37, 708–729. [Google Scholar] [CrossRef]
- Burmistrz, M.; Krakowski, K.; Krawczyk-Balska, A. RNA-targeting CRISPR–Cas systems and their applications. Int. J. Mol. Sci. 2020, 21, 1122. [Google Scholar] [CrossRef]
- Granados-Riveron, J.T.; Aquino-Jarquin, G. CRISPR–Cas13 precision transcriptome engineering in cancer. Cancer Res. 2018, 78, 4107–4113. [Google Scholar] [CrossRef]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1. [Google Scholar] [CrossRef]
- Aman, R.; Mahas, A.; Butt, H.; Ali, Z.; Aljedaani, F.; Mahfouz, M. Engineering RNA virus interference via the CRISPR/Cas13 machinery in Arabidopsis. Viruses 2018, 10, 732. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.; Zhang, F.; Zhong, Z.; Chen, R.; Wang, Y.; Chang, L.; Bock, R.; Nie, B.; Zhang, J. Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. 2019, 17, 1814–1822. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, C.; Chen, B.; An, H.; Jiao, Y.; Zhang, F.; Yang, X. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019, 17, 1185. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Pan, Z.; Wang, X.; Bian, X.; Wang, W.; Liang, Q.; Kou, M.; Ji, H.; Li, Y.; Ma, D. Targeting of SPCSV-RNase3 via CRISPR-Cas13 confers resistance against sweet potato virus disease. Mol. Plant Pathol. 2022, 23, 104–117. [Google Scholar] [CrossRef]
- Kushawah, G.; Hernandez-Huertas, L.; Del Prado, J.A.-N.; Martinez-Morales, J.R.; DeVore, M.L.; Hassan, H.; Moreno-Sanchez, I.; Tomas-Gallardo, L.; Diaz-Moscoso, A.; Monges, D.E. CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos. Dev. Cell 2020, 54, 805–817. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Xue, W.; Zhang, L.; Yang, L.-Z.; Cao, S.-M.; Lei, Y.-N.; Liu, C.-X.; Guo, S.-K.; Shan, L. Screening for functional circular RNAs using the CRISPR–Cas13 system. Nat. Methods 2021, 18, 51–59. [Google Scholar] [CrossRef]
- Ishola, A.A.; Chien, C.-S.; Yang, Y.-P.; Chien, Y.; Yarmishyn, A.A.; Tsai, P.-H.; Chen, J.C.-Y.; Hsu, P.-K.; Luo, Y.-H.; Chen, Y.-M. Oncogenic circRNA hsa_circ_0000190 modulates EGFR/ERK pathway in promoting NSCLC. Cancer Res. 2021, 82, 75–89. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, J.; Zhu, Z.; Zhu, Z.; Liao, X.; Wu, J.; Cheng, J.; Zhang, X.; Mei, H.; Yang, G. CRISPR-Cas13-mediated knockdown of lncRNA-GACAT3 inhibited cell proliferation and motility, and induced apoptosis by increasing p21, Bax, and E-cadherin expression in bladder cancer. Front. Mol. Biosci. 2021, 7, 433. [Google Scholar] [CrossRef]
- Xu, D.; Cai, Y.; Tang, L.; Han, X.; Gao, F.; Cao, H.; Qi, F.; Kapranov, P. A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci. Rep. 2020, 10, 1794. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Franklin, B.; Koob, J.; Kellner, M.J.; Ladha, A.; Joung, J.; Kirchgatterer, P.; Cox, D.B.; Zhang, F. A cytosine deaminase for programmable single-base RNA editing. Science 2019, 365, 382–386. [Google Scholar] [CrossRef]
- Kannan, S.; Altae-Tran, H.; Jin, X.; Madigan, V.J.; Oshiro, R.; Makarova, K.S.; Koonin, E.V.; Zhang, F. Compact RNA editors with small Cas13 proteins. Nat. Biotechnol. 2022, 40, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.-L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef]
- Kang, B.-C.; Yun, J.-Y.; Kim, S.-T.; Shin, Y.; Ryu, J.; Choi, M.; Woo, J.W.; Kim, J.-S. Precision genome engineering through adenine base editing in plants. Nat. Plants 2018, 4, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Tao, X.; Yuan, F.; Wang, D.; Zhu, J.-K. Precise A· T to G· C base editing in the rice genome. Mol. Plant 2018, 11, 627–630. [Google Scholar] [CrossRef]
- Li, J.; Sun, Y.; Du, J.; Zhao, Y.; Xia, L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant 2017, 10, 526–529. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y. Precise plant genome editing using base editors and prime editors. Nat. Plants 2021, 7, 1166–1187. [Google Scholar] [CrossRef]
- Morton, M.; AlTamimi, N.; Butt, H.; Reddy, A.S.; Mahfouz, M. Serine/Arginine-rich protein family of splicing regulators: New approaches to study splice isoform functions. Plant Sci. 2019, 283, 127–134. [Google Scholar] [CrossRef]
- Yang, L.-Z.; Wang, Y.; Li, S.-Q.; Yao, R.-W.; Luan, P.-F.; Wu, H.; Carmichael, G.G.; Chen, L.-L. Dynamic imaging of RNA in living cells by CRISPR-Cas13 systems. Mol. Cell 2019, 76, 981–997. [Google Scholar] [CrossRef]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Simon, H.Y.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cao, J.; Xiong, M.; Petersen, A.J.; Dong, Y.; Tao, Y.; Huang, C.T.-L.; Du, Z.; Zhang, S.-C. Engineering human stem cell lines with inducible gene knockout using CRISPR/Cas9. Cell Stem Cell 2015, 17, 233–244. [Google Scholar] [CrossRef]
- Jia, Y.; Xu, R.-G.; Ren, X.; Ewen-Campen, B.; Rajakumar, R.; Zirin, J.; Yang-Zhou, D.; Zhu, R.; Wang, F.; Mao, D. Next-generation CRISPR/Cas9 transcriptional activation in Drosophila using flySAM. Proc. Natl. Acad. Sci. USA 2018, 115, 4719–4724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qin, W.; Lu, X.; Xu, J.; Huang, H.; Bai, H.; Li, S.; Lin, S. Programmable base editing of zebrafish genome using a modified CRISPR-Cas9 system. Nat. Commun. 2017, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, D.; Gutkin, A.; Kedmi, R.; Ramishetti, S.; Veiga, N.; Jacobi, A.M.; Schubert, M.S.; Friedmann-Morvinski, D.; Cohen, Z.R.; Behlke, M.A. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 2020, 6, eabc9450. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Hu, L.; Ying, L.; Zhao, Z.; Chu, P.K.; Yu, X.-F. A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018, 9, 5012. [Google Scholar] [CrossRef]
- Li, R.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 2018, 94, 513–524. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, K.; Cheng, Y.; Zhong, Z.; Tian, L.; Tang, X.; Tang, A.; Zheng, X.; Zhang, T.; Qi, Y. CRISPR-Cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 2017, 8, 1598. [Google Scholar] [CrossRef]
- Zhou, J.; Yuan, M.; Zhao, Y.; Quan, Q.; Yu, D.; Yang, H.; Tang, X.; Xin, X.; Cai, G.; Qian, Q. Efficient deletion of multiple circle RNA loci by CRISPR-Cas9 reveals Os06circ02797 as a putative sponge for OsMIR408 in rice. Plant Biotechnol. 2021, 19, 1240. [Google Scholar] [CrossRef]
- Veillet, F.; Perrot, L.; Chauvin, L.; Kermarrec, M.-P.; Guyon-Debast, A.; Chauvin, J.-E.; Nogué, F.; Mazier, M. Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 2019, 20, 402. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Li, J.; Wang, Q.; Xu, Z.; Sun, L.; Alariqi, M.; Manghwar, H.; Wang, G.; Li, B.; Ding, X. High-efficient and precise base editing of C• G to T• A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol. 2020, 18, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Chester, A.; Scott, J.; Anant, S.; Navaratnam, N. RNA editing: Cytidine to uridine conversion in apolipoprotein B mRNA. Biochim. Biophys. Acta Gene Struct. Expr. 2000, 1494, 1–13. [Google Scholar] [CrossRef]
- Bharat, S.S.; Li, S.; Li, J.; Yan, L.; Xia, L. Base editing in plants: Current status and challenges. Crop J. 2020, 8, 384–395. [Google Scholar] [CrossRef]
- Mishra, R.; Joshi, R.K.; Zhao, K. Base editing in crops: Current advances, limitations and future implications. Plant Biotechnol. 2020, 18, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, X.; Zhou, J.; Yang, C.; Wang, G.; Tan, Y.; Wu, Y.; Zhang, S.; Yi, K.; Kang, C. The CRISPR-cas13a gene-editing system induces collateral cleavage of RNA in glioma cells. Adv. Sci. 2019, 6, 1901299. [Google Scholar] [CrossRef]
- Buchman, A.B.; Brogan, D.J.; Sun, R.; Yang, T.; Hsu, P.D.; Akbari, O.S. Programmable RNA targeting using CasRx in flies. CRISPR J. 2020, 3, 164–176. [Google Scholar] [CrossRef]
- Li, Y.; Xu, J.; Guo, X.; Li, Z.; Cao, L.; Liu, S.; Guo, Y.; Wang, G.; Luo, Y.; Zhang, Z. Collateral cleavage of 28s rRNA by RfxCas13d causes death of mice. bioRxiv 2022. [Google Scholar] [CrossRef]
- Tong, H.; Huang, J.; Xiao, Q.; He, B.; Dong, X.; Liu, Y.; Yang, X.; Han, D.; Wang, Z.; Wang, X. High-fidelity Cas13 variants for targeted RNA degradation with minimal collateral effects. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Tng, P.Y.L.; Carabajal Paladino, L.; Verkuijl, S.A.N.; Purcell, J.; Merits, A.; Leftwich, P.T.; Fragkoudis, R.; Noad, R.; Alphey, L. Cas13b-dependent and Cas13b-independent RNA knockdown of viral sequences in mosquito cells following guide RNA expression. Commun. Biol. 2020, 3, 413. [Google Scholar] [CrossRef]
- Wu, Q.-W.; Kapfhammer, J.P. The bacterial enzyme Cas13 interferes with neurite outgrowth from cultured cortical neurons. Toxins 2021, 13, 262. [Google Scholar] [CrossRef] [PubMed]
- Charles, E.J.; Kim, S.E.; Knott, G.J.; Smock, D.; Doudna, J.; Savage, D.F. Engineering improved Cas13 effectors for targeted post-transcriptional regulation of gene expression. bioRxiv 2021. [Google Scholar] [CrossRef]

| Application Scope | Cas13 Type | Plant Species | Target | Reference |
|---|---|---|---|---|
| Viral RNA interference | LshCas13a | Nicotiana benthamiana | Turnip Mosaic Virus (TuMV) | [40] |
| Arabidopsis thaliana | [41] | |||
| Solanum tuberosum | Potato Virus Y (PVY) | [42] | ||
| Nicotiana benthamiana, Oryza sativa | Southern Rice Black-Streaked Dwarf Virus (SRBSDV), Rice Stripe Mosaic Virus (RSMV) | [43] | ||
| LshCas13a, LwaCas13a, PspCas13b, BzCas13b, RfxCas13d | Nicotiana benthamiana | Turnip Mosaic Virus (TuMV), Tobacco Mosaic Virus (TMV), Potato Virus X (PVX) | [30] | |
| LshCas13a, LwaCas13a, PspCas13b, RfxCas13d | Nicotiana benthamiana, Ipomoea batatas | Turnip Mosaic Virus (TuMV), Cucumber Mosaic Virus (CMV), Sweet Potato Chlorotic Stunt Virus (SPCSV)-RNase3 | [44] | |
| mRNA knockdown | LwaCas13a | Oryza sativa ssp. Japonica var. Nipponbare (Protoplasts) | 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), hydroxycinnamoyl transferase (HCT), and phytoene desaturase (PDS) | [25] |
| LbaCas13a, LbuCas13a | Nicotiana benthamiana, Arabidopsis thaliana, Solanum lycopersicum | PDS transcript | [15] | |
| RNA detection | LwaCas13a, PsmCas13b | (Glyphosate resistant) Glycine max | EPSPS from Agrobacterium sp. strain CP4 (CP4 EPSPS) | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kavuri, N.R.; Ramasamy, M.; Qi, Y.; Mandadi, K. Applications of CRISPR/Cas13-Based RNA Editing in Plants. Cells 2022, 11, 2665. https://doi.org/10.3390/cells11172665
Kavuri NR, Ramasamy M, Qi Y, Mandadi K. Applications of CRISPR/Cas13-Based RNA Editing in Plants. Cells. 2022; 11(17):2665. https://doi.org/10.3390/cells11172665
Chicago/Turabian StyleKavuri, Naga Rajitha, Manikandan Ramasamy, Yiping Qi, and Kranthi Mandadi. 2022. "Applications of CRISPR/Cas13-Based RNA Editing in Plants" Cells 11, no. 17: 2665. https://doi.org/10.3390/cells11172665
APA StyleKavuri, N. R., Ramasamy, M., Qi, Y., & Mandadi, K. (2022). Applications of CRISPR/Cas13-Based RNA Editing in Plants. Cells, 11(17), 2665. https://doi.org/10.3390/cells11172665