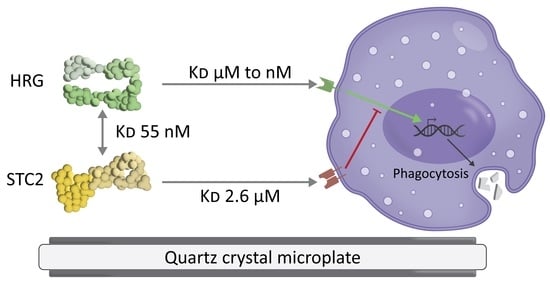
Quartz Crystal Microbalance Measurement of Histidine-Rich Glycoprotein and Stanniocalcin-2 Binding to Each Other and to Inflammatory Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Differentiation of U937 Cells
2.2. Purified Proteins and Phagocytosis Assay
2.3. Co-Immunoprecipitation and Immunoblotting
2.4. Binding of HRG to STC2
2.5. Binding of HRG and STC2 to U937 Cells Treated or Not with Heparinase and Fixative
3. Results
3.1. HRG Increases Phagocytosis by U937 Monocytes
3.2. HRG and STC2 Interact with Nanomolar Affinity
3.3. Live U937 Cells Bind HRG after vitD3 Differentiation
3.4. Binding of HRG to Fixed U937 Cells Is Independent of vitD3-Induced Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Rolny, C.; Mazzone, M.; Tugues, S.; Laoui, D.; Johansson, I.; Coulon, C.; Squadrito, M.L.; Segura, I.; Li, X.; Knevels, E.; et al. HRG inhibits tumor growth and metastasis by inducing macrophage polarization and vessel normalization through downregulation of PlGF. Cancer Cell 2011, 19, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, S. Chapter Nine—New Insights into the Functions of Histidine-Rich Glycoprotein. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 467–493. [Google Scholar]
- Poon, I.K.H.; Patel, K.K.; Davis, D.S.; Parish, C.R.; Hulett, M.D. Histidine-rich glycoprotein: The Swiss Army knife of mammalian plasma. Blood 2011, 117, 2093–2101. [Google Scholar] [CrossRef] [PubMed]
- Shannon, O.; Rydengård, V.; Schmidtchen, A.; Mörgelin, M.; Alm, P.; Sørensen, O.E.; Björck, L. Histidine-rich glycoprotein promotes bacterial entrapment in clots and decreases mortality in a mouse model of sepsis. Blood 2010, 116, 2365–2372. [Google Scholar] [CrossRef]
- Gorgani, N.N.; Parish, C.; Easterbrook Smith, S.B.; Altin, J.G. Histidine-Rich Glycoprotein Binds to Human IgG and C1q and Inhibits the Formation of Insoluble Immune Complexes. Biochemistry 1997, 36, 6653–6662. [Google Scholar] [CrossRef] [PubMed]
- Vanwildemeersch, M.; Olsson, A.-K.; Gottfridsson, E.; Claesson-Welsh, L.; Lindahl, U.; Spillmann, D. The anti-angiogenic His/Pro-rich fragment of histidine-rich glycoprotein binds to endothelial cell heparan sulfate in a Zn2+-dependent manner. J. Biol. Chem. 2006, 281, 10298–10304. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.-K.; Larsson, H.; Dixelius, J.; Johansson, I.; Lee, C.; Oellig, C.; Björk, I.; Claesson-Welsh, L. A fragment of histidine-rich glycoprotein is a potent inhibitor of tumor vascularization. Cancer Res. 2004, 64, 599–605. [Google Scholar] [CrossRef]
- Kassaar, O.; McMahon, S.A.; Thompson, R.; Botting, C.H.; Naismith, J.H.; Stewart, A.J. Crystal structure of histidine-rich glycoprotein N2 domain reveals redox activity at an interdomain disulfide bridge: Implications for angiogenic regulation. Blood 2014, 123, 1948–1955. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Blank, M.; Shoenfeld, Y. Histidine-rich glycoprotein modulation of immune/autoimmune, vascular, and coagulation systems. Clin. Rev. Allergy Immunol. 2008, 34, 307–312. [Google Scholar] [CrossRef]
- Roche, F.; Sipilä, K.; Honjo, S.; Johansson, S.; Tugues, S.; Heino, J.; Claesson-Welsh, L. Histidine-rich glycoprotein blocks collagen-binding integrins and adhesion of endothelial cells through low-affinity interaction with alpha2 integrin. Matrix Biol. 2015, 48, 89–99. [Google Scholar] [CrossRef]
- Kuroda, K.; Ishii, K.; Mihara, Y.; Kawanoue, N.; Wake, H.; Mori, S.; Yoshida, M.; Nishibori, M.; Morimatsu, H. Histidine-rich glycoprotein as a prognostic biomarker for sepsis. Sci. Rep. 2021, 11, 10223. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida-Straeten, N.; Ensslen, S.; Schäfer, C.; Wöltje, M.; Denecke, B.; Moser, M.; Gräber, S.; Wakabayashi, S.; Koide, T.; Jahnen-Dechent, W. Enhanced blood coagulation and fibrinolysis in mice lacking histidine-rich glycoprotein (HRG). J. Thromb. Haemost. 2005, 3, 865–872. [Google Scholar] [CrossRef]
- Lee, C.; Bongcam-Rudloff, E.; Sollner, C.; Jahnen-Dechent, W.; Claesson-Welsh, L. Type 3 cystatins; fetuins, kininogen and histidine-rich glycoprotein. Front. Biosci. 2009, 14, 2911–2922. [Google Scholar] [CrossRef] [PubMed]
- Tugues, S.; Honjo, S.; König, C.; Noguer, O.; Hedlund, M.; Botling, J.; Deschoemaeker, S.; Wenes, M.; Rolny, C.; Jahnen-Dechent, W.; et al. Genetic deficiency in plasma protein HRG enhances tumor growth and metastasis by exacerbating immune escape and vessel abnormalization. Cancer Res. 2012, 72, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Roche, F.P.; Pietilä, I.; Kaito, H.; Sjöström, E.O.; Sobotzki, N.; Noguer, O.; Skare, T.P.; Essand, M.; Wollscheid, B.; Welsh, M.; et al. Leukocyte Differentiation by Histidine-Rich Glycoprotein/Stanniocalcin-2 Complex Regulates Murine Glioma Growth through Modulation of Antitumor Immunity. Mol. Cancer Ther. 2018, 17, 1961–1972. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Joshi, A.D. New Insights Into Physiological and Pathophysiological Functions of Stanniocalcin 2. Front. Endocrinol. 2020, 11, 172. [Google Scholar] [CrossRef]
- Yeung, B.H.; Law, A.Y.; Wong, C.K. Evolution and roles of stanniocalcin. Mol. Cell Endocrinol. 2012, 349, 272–280. [Google Scholar] [CrossRef]
- Chang, A.C.; Hook, J.; Lemckert, F.A.; McDonald, M.; Nguyen, M.-A.T.; Hardeman, E.C.; Little, D.G.; Gunning, P.W.; Reddel, R. The murine stanniocalcin 2 gene is a negative regulator of postnatal growth. Endocrinology 2008, 149, 2403–2410. [Google Scholar] [CrossRef]
- Gianneli, M.; Polo, E.; Lopez, H.; Castagnola, V.; Aastrup, T.; Dawson, K.A. Label-free in-flow detection of receptor recognition motifs on the biomolecular corona of nanoparticles. Nanoscale 2018, 10, 5474–5481. [Google Scholar] [CrossRef]
- Olsson, I.; Gullberg, U.; Ivhed, I.; Nilsson, K. Induction of differentiation of the human histiocytic lymphoma cell line U-937 by 1 alpha,25-dihydroxycholecalciferol. Cancer Res. 1983, 43, 5862–5867. [Google Scholar] [PubMed]
- Sundström, C.; Nilsson, K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int. J. Cancer 1976, 17, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Tugues, S.; Roche, F.; Noguer, O.; Orlova, A.; Bhoi, S.; Padhan, N.; Åkerud, P.; Honjo, S.; Selvaraju, R.K.; Mazzone, M.; et al. Histidine-rich glycoprotein uptake and turnover is mediated by mononuclear phagocytes. PLoS ONE 2014, 9, e107483. [Google Scholar]
- Peiris, D.; Aastrup, T.; Altun, S.; Käck, C.; Gianneli, M.; Proverbio, D.; Jørgensen, L.M. Label-Free Cell-Based Assay for Characterization of Biomolecules and Receptors. In Epitope Mapping Protocols; Rockberg, J., Nilvebrant, J., Eds.; Springer: New York, NY, USA, 2018; pp. 53–63. [Google Scholar]
- Forssén, P.; Multia, E.; Samuelsson, J.; Andersson, M.; Aastrup, T.; Altun, S.; Wallinder, D.; Wallbing, L.; Liangsupree, T.; Riekkola, M.-L.; et al. Reliable Strategy for Analysis of Complex Biosensor Data. Anal. Chem. 2018, 90, 5366–5374. [Google Scholar] [CrossRef] [PubMed]
- Theek, B.; Baues, M.; Gremse, F.; Pola, R.; Pechar, M.; Negwer, I.; Koynov, K.; Weber, B.; Barz, M.; Jahnen-Dechent, W.; et al. Histidine-rich glycoprotein-induced vascular normalization improves EPR-mediated drug targeting to and into tumors. J. Control. Release 2018, 282, 25–34. [Google Scholar] [CrossRef]
- Marx, K.A. Quartz Crystal Microbalance: A Useful Tool for Studying Thin Polymer Films and Complex Biomolecular Systems at the Solution−Surface Interface. Biomacromolecules 2003, 4, 1099–1120. [Google Scholar] [CrossRef]
- Pei, Z.; Saint-Guirons, J.; Käck, C.; Ingemarsson, B.; Aastrup, T. Real-time analysis of the carbohydrates on cell surfaces using a QCM biosensor: A lectin-based approach. Biosens. Bioelectron. 2012, 35, 200–205. [Google Scholar] [CrossRef]
- Paavilainen, L.; Edvinsson, Å.; Asplund, A.; Hober, S.; Kampf, C.; Pontén, F.; Wester, K. The impact of tissue fixatives on morphology and antibody-based protein profiling in tissues and cells. J. Histochem. Cytochem. 2010, 58, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Zamani, F.; Shahneh, F.Z.; Aghebati-Maleki, L.; Baradaran, B. Induction of CD14 Expression and Differentiation to Monocytes or Mature Macrophages in Promyelocytic Cell Lines: New Approach. Adv. Pharm. Bull. 2013, 3, 329–332. [Google Scholar]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Persson Skare, T.; Kaito, H.; Durall, C.; Aastrup, T.; Claesson-Welsh, L. Quartz Crystal Microbalance Measurement of Histidine-Rich Glycoprotein and Stanniocalcin-2 Binding to Each Other and to Inflammatory Cells. Cells 2022, 11, 2684. https://doi.org/10.3390/cells11172684
Persson Skare T, Kaito H, Durall C, Aastrup T, Claesson-Welsh L. Quartz Crystal Microbalance Measurement of Histidine-Rich Glycoprotein and Stanniocalcin-2 Binding to Each Other and to Inflammatory Cells. Cells. 2022; 11(17):2684. https://doi.org/10.3390/cells11172684
Chicago/Turabian StylePersson Skare, Tor, Hiroshi Kaito, Claudia Durall, Teodor Aastrup, and Lena Claesson-Welsh. 2022. "Quartz Crystal Microbalance Measurement of Histidine-Rich Glycoprotein and Stanniocalcin-2 Binding to Each Other and to Inflammatory Cells" Cells 11, no. 17: 2684. https://doi.org/10.3390/cells11172684
APA StylePersson Skare, T., Kaito, H., Durall, C., Aastrup, T., & Claesson-Welsh, L. (2022). Quartz Crystal Microbalance Measurement of Histidine-Rich Glycoprotein and Stanniocalcin-2 Binding to Each Other and to Inflammatory Cells. Cells, 11(17), 2684. https://doi.org/10.3390/cells11172684