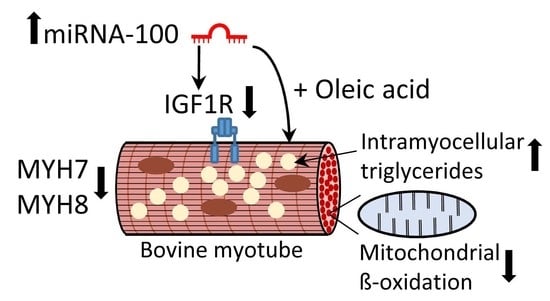
MicroRNA-100 Reduced Fetal Bovine Muscle Satellite Cell Myogenesis and Augmented Intramuscular Lipid Deposition by Modulating IGF1R
Abstract
:1. Introduction
2. Materials and Methods
2.1. MicroRNA Target Prediction and Dual-Luciferase Reporter Assay
2.2. Isolation of Bovine Muscle Satellite Cells
2.3. Muscle Satellite Cell Culture and miRNA Transfection
2.4. RNA Extraction and Real-Time PCR
2.5. Protein Extraction and Western Blotting
2.6. Detection of miR-100 Expression
2.7. Oil Red O Staining
2.8. BrdU (5-bromo-2′-deoxyuridine) Labelling of Myoblasts
2.9. Immunocytochemistry
2.10. Statistical Analyses
3. Results
3.1. MiR-100 Directly Targets Bovine IGF1R
3.2. Correlative Expression of miR-100 and IGF1R in Bovine Muscle Satellite Cell Myogenesis
3.3. MiR-100 Modulates IGF1R/PI3K/AKT Signaling through IGF1R Suppression
3.4. IGF1R Inhibition Has No Impact on miR-100 Levels
3.5. MiR-100 Did Not Influence Proliferation of BMSC
3.6. Overexpression of miR-100 Promotes Oleic Acid-Induced Lipid Deposition and Reduces Lipid Oxidation in Myotubes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cossu, G.; Borello, U. Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999, 18, 6867–6872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stickland, N.C. A quantitative study of muscle development in the bovine foetus (Bos indicus). Anat. Histol. Embryol. 1978, 7, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Tong, J.; Zhao, J.; Underwood, K.R.; Zhu, M.; Ford, S.P.; Nathanielsz, P.W. Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 2010, 88 (Suppl. 13), E51–E60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, P.; Holowacz, T.; Lassar, A.B. The origin of skeletal muscle stem cells in the embryo and the adult. Curr. Opin. Cell Biol. 2001, 13, 679–689. [Google Scholar] [CrossRef]
- Cristancho, A.G.; Lazar, M.A. Forming functional fat: A growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell Biol. 2011, 12, 722–734. [Google Scholar] [CrossRef]
- Tong, J.F.; Yan, X.; Zhu, M.J.; Ford, S.P.; Nathanielsz, P.W.; Du, M. Maternal obesity downregulates myogenesis and beta-catenin signaling in fetal skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E917–E924. [Google Scholar] [CrossRef] [Green Version]
- Vettor, R.; Milan, G.; Franzin, C.; Sanna, M.; De Coppi, P.; Rizzuto, R.; Federspil, G. The origin of intermuscular adipose tissue and its pathophysiological implications. Am. J. Physiol. Endocrinol. Metab. 2009, 297, E987–E998. [Google Scholar] [CrossRef]
- Bakke, S.S.; Moro, C.; Nikolić, N.; Hessvik, N.P.; Badin, P.M.; Lauvhaug, L.; Fredriksson, K.; Hesselink, M.K.; Boekschoten, M.V.; Kersten, S.; et al. Palmitic acid follows a different metabolic pathway than oleic acid in human skeletal muscle cells; lower lipolysis rate despite an increased level of adipose triglyceride lipase. Biochim. Biophys. Acta 2012, 1821, 1323–1333. [Google Scholar] [CrossRef]
- Xiang, A.; Chu, G.; Zhu, Y.; Ma, G.; Yang, G.; Sun, S. IGFBP5 suppresses oleate-induced intramyocellular lipids deposition and enhances insulin signaling. J. Cell Physiol. 2019, 234, 15288–15298. [Google Scholar] [CrossRef]
- Chargé, S.B.; Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef]
- Florini, J.R.; Ewton, D.Z.; Coolican, S.A. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr. Rev. 1996, 17, 481–517. [Google Scholar] [CrossRef] [Green Version]
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.; Liu, J.P.; Robertson, E.J.; Efstratiadis, A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 1993, 75, 73–82. [Google Scholar] [CrossRef]
- Glass, D.J. Skeletal muscle hypertrophy and atrophy signaling pathways. Int. J. Biochem. Cell Biol. 2005, 37, 1974–1984. [Google Scholar] [CrossRef] [PubMed]
- Rommel, C.; Bodine, S.C.; Clarke, B.A.; Rossman, R.; Nunez, L.; Stitt, T.N.; Yancopoulos, G.D.; Glass, D.J. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001, 3, 1009–1013. [Google Scholar] [CrossRef]
- Bassel-Duby, R.; Olson, E.N. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 2006, 75, 19–37. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, S.S.; Ahmad, K.; Lee, E.J.; Lee, Y.-H.; Choi, I. Implications of Insulin-Like Growth Factor-1 in Skeletal Muscle and Various Diseases. Cells 2020, 9, 1773. [Google Scholar] [CrossRef]
- Xu, Q.; Wu, Z. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J. Biol. Chem. 2000, 275, 36750–36757. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.P.; Baker, J.; Perkins, A.S.; Robertson, E.J.; Efstratiadis, A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 1993, 75, 59–72. [Google Scholar] [CrossRef]
- Mavalli, M.D.; DiGirolamo, D.J.; Fan, Y.; Riddle, R.C.; Campbell, K.S.; van Groen, T.; Frank, S.J.; Sperling, M.A.; Esser, K.A.; Bamman, M.M.; et al. Distinct growth hormone receptor signaling modes regulate skeletal muscle development and insulin sensitivity in mice. J. Clin. Investig. 2010, 120, 4007–4020. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, B.T.; Lauritzen, H.P.; Hirshman, M.F.; Smyth, G.; Goodyear, L.J.; Kahn, C.R. Differential Role of Insulin/IGF-1 Receptor Signaling in Muscle Growth and Glucose Homeostasis. Cell Rep. 2015, 11, 1220–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, M.E.; DeMayo, F.; Yin, K.C.; Lee, H.M.; Geske, R.; Montgomery, C.; Schwartz, R.J. Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. J. Biol. Chem. 1995, 270, 12109–12116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, L.S.; Anderson, B.G.; Plymate, S.R. Muscle-specific overexpression of the type 1 IGF receptor results in myoblast-independent muscle hypertrophy via PI3K, and not calcineurin, signaling. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E1538–E1551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klöting, N.; Koch, L.; Wunderlich, T.; Kern, M.; Ruschke, K.; Krone, W.; Brüning, J.C.; Blüher, M. Autocrine IGF-1 action in adipocytes controls systemic IGF-1 concentrations and growth. Diabetes 2008, 57, 2074–2082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boucher, J.; Softic, S.; El Ouaamari, A.; Krumpoch, M.T.; Kleinridders, A.; Kulkarni, R.N.; O’Neill, B.T.; Kahn, C.R. Differential Roles of Insulin and IGF-1 Receptors in Adipose Tissue Development and Function. Diabetes 2016, 65, 2201–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Sayed, D.; Abdellatif, M. MicroRNAs in Development and Disease. Physiol. Rev. 2011, 91, 827–887. [Google Scholar] [CrossRef]
- Wang, J.; Yang, L.Z.; Zhang, J.S.; Gong, J.X.; Wang, Y.H.; Zhang, C.L.; Chen, H.; Fang, X.T. Effects of microRNAs on skeletal muscle development. Gene 2018, 668, 107–113. [Google Scholar] [CrossRef]
- Singh, G.B.; Cowan, D.B.; Wang, D.-Z. Tiny Regulators of Massive Tissue: MicroRNAs in Skeletal Muscle Development, Myopathies, and Cancer Cachexia. Front. Oncol. 2020, 10, 598964. [Google Scholar] [CrossRef]
- Mir, B.A.; Islam, R.; Kalanon, M.; Russell, A.P.; Foletta, V.C. MicroRNA suppression of stress-responsive NDRG2 during dexamethasone treatment in skeletal muscle cells. BMC Mol. Cell Biol. 2019, 20, 12. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Qian, K.; Wang, C. Discovery of porcine miRNA-196a/b may influence porcine adipogenesis in longissimus dorsi muscle by miRNA sequencing. Anim. Genet. 2017, 48, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.F.; Xiong, Y.; Peng, Y.; Gao, Y.; Qin, J.; Chu, G.Y.; Pang, W.J.; Yang, G.S. miR-425-5p Inhibits Differentiation and Proliferation in Porcine Intramuscular Preadipocytes. Int. J. Mol. Sci. 2017, 18, 2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Wang, S.; Feng, X.; Liu, X.; Zhao, J.; Zheng, Q.; Wei, X.; Ma, Y. miRNA transcriptome comparison between muscle and adipose tissues indicates potential miRNAs associated with intramuscular fat in Chinese swamp buffalo. Genome 2019, 62, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, Q.-S.; Li, W.-B.; Wei, J.; Chang, W.-K.; Chen, Z.; Qiao, H.-Y.; Jia, Y.-W.; Tian, J.-H.; Liang, B.-S. miRNA targeted signaling pathway in the early stage of denervated fast and slow muscle atrophy. Neural Regen. Res. 2016, 11, 1293–1303. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Martello, G.; Piccolo, S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010, 11, 252–263. [Google Scholar] [CrossRef]
- Wu, Y.; Li, H.; Xie, J.; Wang, F.; Cao, D.; Lou, Y. miR-139-5p affects cell proliferation, migration and adipogenesis by targeting insulin-like growth factor 1 receptor in hemangioma stem cells. Int. J. Mol. Med. 2020, 45, 569–577. [Google Scholar] [CrossRef] [Green Version]
- Das, F.; Dey, N.; Bera, A.; Kasinath, B.S.; Ghosh-Choudhury, N.; Choudhury, G.G. MicroRNA-214 Reduces Insulin-like Growth Factor-1 (IGF-1) Receptor Expression and Downstream mTORC1 Signaling in Renal Carcinoma Cells. J. Biol. Chem. 2016, 291, 14662–14676. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Wu, J.; Li, Z.; Qin, H.; Wang, B.; Wong, T.-S.; Yang, W.; Fu, Q.-L.; Lei, W. miR-375 Suppresses IGF1R Expression and Contributes to Inhibition of Cell Progression in Laryngeal Squamous Cell Carcinoma. Biomed. Res. Int. 2014, 2014, 374598. [Google Scholar] [CrossRef]
- Knezevic, I.; Patel, A.; Sundaresan, N.R.; Gupta, M.P.; Solaro, R.J.; Nagalingam, R.S.; Gupta, M. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: Implications in postnatal cardiac remodeling and cell survival. J. Biol. Chem. 2012, 287, 12913–12926. [Google Scholar] [CrossRef] [Green Version]
- Pek, S.L.T.; Sum, C.F.; Lin, M.X.; Cheng, A.K.S.; Wong, M.T.K.; Lim, S.C.; Tavintharan, S. Circulating and visceral adipose miR-100 is down-regulated in patients with obesity and Type 2 diabetes. Mol. Cell. Endocrinol. 2016, 427, 112–123. [Google Scholar] [CrossRef]
- Huang, M.B.; Xu, H.; Xie, S.J.; Zhou, H.; Qu, L.H. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS ONE 2011, 6, e29173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, S.; Wassler, M.; Zhang, L.; Li, Y.; Wang, J.; Zhang, Y.; Shelat, H.; Williams, J.; Geng, Y.-J. MicroRNA-133a regulates insulin-like growth factor-1 receptor expression and vascular smooth muscle cell proliferation in murine atherosclerosis. Atherosclerosis 2014, 232, 171–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tewari, R.S.; Ala, U.; Accornero, P.; Baratta, M.; Miretti, S. Circulating skeletal muscle related microRNAs profile in Piedmontese cattle during different age. Sci. Rep. 2021, 11, 15815. [Google Scholar] [CrossRef] [PubMed]
- Sylvius, N.; Bonne, G.; Straatman, K.; Reddy, T.; Gant, T.W.; Shackleton, S. MicroRNA expression profiling in patients with lamin A/C-associated muscular dystrophy. FASEB J. 2011, 25, 3966–3978. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, P.; Stankevicins, L.; Ansseau, E.; Petrov, A.; Barat, A.; Dessen, P.; Robert, T.; Turki, A.; Lazar, V.; Labourer, E.; et al. Defective regulation of microRNA target genes in myoblasts from facioscapulohumeral dystrophy patients. J. Biol. Chem. 2013, 288, 34989–35002. [Google Scholar] [CrossRef] [Green Version]
- Margolis, L.M.; Berryman, C.E.; Murphy, N.E.; Carrigan, C.T.; Young, A.J.; Carbone, J.W.; Pasiakos, S.M. PI3K-AKT-FOXO1 pathway targeted by skeletal muscle microRNA to suppress proteolytic gene expression in response to carbohydrate intake during aerobic exercise. Physiol. Rep. 2018, 6, e13931. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, C.J.; D’Souza, R.F.; Schierding, W.; Zeng, N.; Ramzan, F.; O’Sullivan, J.M.; Poppitt, S.D.; Cameron-Smith, D. Identification of human skeletal muscle miRNA related to strength by high-throughput sequencing. Physiol. Genom. 2018, 50, 416–424. [Google Scholar] [CrossRef]
- de Oliveira, P.S.N.; Coutinho, L.L.; Cesar, A.S.M.; Diniz, W.; de Souza, M.M.; Andrade, B.G.; Koltes, J.E.; Mourão, G.B.; Zerlotini, A.; Reecy, J.M.; et al. Co-Expression Networks Reveal Potential Regulatory Roles of miRNAs in Fatty Acid Composition of Nelore Cattle. Front. Genet. 2019, 10, 651. [Google Scholar] [CrossRef] [Green Version]
- Mir, B.A.; Reyer, H.; Komolka, K.; Ponsuksili, S.; Kühn, C.; Maak, S. Differentially Expressed miRNA-Gene Targets Related to Intramuscular Fat in Musculus Longissimus Dorsi of Charolais × Holstein F2-Crossbred Bulls. Genes 2020, 11, 700. [Google Scholar] [CrossRef]
- Komolka, K.; Ponsuksili, S.; Albrecht, E.; Kühn, C.; Wimmers, K.; Maak, S. Gene expression profile of Musculus longissimus dorsi in bulls of a Charolais × Holstein F2-cross with divergent intramuscular fat content. Genom. Data 2016, 7, 131–133. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- ThermoFisher, New Tools for MicroRNA Functional Analysis, mirVana™ miRNA Mimics and Inhibitors. BioProbes, J. Cell Biol. Appl. 2012, 67, 20–21.
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schering, L.; Albrecht, E.; Komolka, K.; Kühn, C.; Maak, S. Increased expression of thyroid hormone responsive protein (THRSP) is the result but not the cause of higher intramuscular fat content in cattle. Int. J. Biol. Sci. 2017, 13, 532–544. [Google Scholar] [CrossRef] [Green Version]
- Dahl, N.; Albrecht, E.; Dannenberger, D.; Uken, K.L.; Hammon, H.M.; Maak, S. Consequences of Maternal Essential Fatty Acid and Conjugated Linoleic Acid Supplementation on the Development of Calf Muscle and Adipose Tissue. Animals 2020, 10, 1598. [Google Scholar] [CrossRef]
- Yu, M.; Wang, H.; Xu, Y.; Yu, D.; Li, D.; Liu, X.; Du, W. Insulin-like growth factor-1 (IGF-1) promotes myoblast proliferation and skeletal muscle growth of embryonic chickens via the PI3K/Akt signalling pathway. Cell Biol. Int. 2015, 39, 910–922. [Google Scholar] [CrossRef]
- Lee, E.K.; Gorospe, M. Minireview: Posttranscriptional regulation of the insulin and insulin-like growth factor systems. Endocrinology 2010, 151, 1403–1408. [Google Scholar] [CrossRef] [Green Version]
- Werner, H.; Maor, S. The insulin-like growth factor-I receptor gene: A downstream target for oncogene and tumor suppressor action. Trends Endocrinol. Metab. 2006, 17, 236–242. [Google Scholar] [CrossRef]
- Jin, Y.; Tymen, S.D.; Chen, D.; Fang, Z.J.; Zhao, Y.; Dragas, D.; Dai, Y.; Marucha, P.T.; Zhou, X. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS ONE 2013, 8, e64434. [Google Scholar] [CrossRef]
- Wei, F.; Liu, Y.; Guo, Y.; Xiang, A.; Wang, G.; Xue, X.; Lu, Z. miR-99b-targeted mTOR induction contributes to irradiation resistance in pancreatic cancer. Mol. Cancer 2013, 12, 81. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Jin, Y.; Yu, D.; Wang, A.; Mahjabeen, I.; Wang, C.; Liu, X.; Zhou, X. Down-regulation of the microRNA-99 family members in head and neck squamous cell carcinoma. Oral Oncol. 2012, 48, 686–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.; Lee, Y.S.; Malhotra, A.; Kim, H.K.; Matecic, M.; Evans, C.; Jensen, R.V.; Moskaluk, C.A.; Dutta, A. miR-99 family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res. 2011, 71, 1313–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Tang, S.; Du, F.; Li, H.; Shen, X.; Li, D.; Wang, Y.; Zhang, Z.; Xia, L.; Zhu, Q.; et al. miR-99a-5p Regulates the Proliferation and Differentiation of Skeletal Muscle Satellite Cells by Targeting MTMR3 in Chicken. Genes 2020, 11, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zacharewicz, E.; Kalanon, M.; Murphy, R.M.; Russell, A.P.; Lamon, S. MicroRNA-99b-5p downregulates protein synthesis in human primary myotubes. Am. J. Physiol. Cell Physiol. 2020, 319, C432–C440. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Huang, Y.; Jiang, C.C.; Zhu, Y.; Wang, L.; Zhang, W.; Huang, W.; Zhou, T.; Tang, S. miR-100 Inhibits Cell Growth and Proliferation by Targeting HOXA1 in Nasopharyngeal Carcinoma. Onco. Targets Ther. 2020, 13, 593–602. [Google Scholar] [CrossRef] [Green Version]
- Elia, L.; Contu, R.; Quintavalle, M.; Varrone, F.; Chimenti, C.; Russo, M.A.; Cimino, V.; Marinis, L.D.; Frustaci, A.; Catalucci, D.; et al. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 2009, 120, 2377–2385. [Google Scholar] [CrossRef]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef]
- Yu, H.; Waddell, J.N.; Kuang, S.; Bidwell, C.A. Park7 expression influences myotube size and myosin expression in muscle. PLoS ONE 2014, 9, e92030. [Google Scholar] [CrossRef]
- Gumucio, J.P.; Qasawa, A.H.; Ferrara, P.J.; Malik, A.N.; Funai, K.; McDonagh, B.; Mendias, C.L. Reduced mitochondrial lipid oxidation leads to fat accumulation in myosteatosis. FASEB J. 2019, 33, 7863–7881. [Google Scholar] [CrossRef]
- van Rooij, E.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J., Jr.; Olson, E.N. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef] [Green Version]
- Mittendorfer, B. Origins of metabolic complications in obesity: Adipose tissue and free fatty acid trafficking. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 535–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Triplett, W.T.; Baligand, C.; Forbes, S.C.; Willcocks, R.J.; Lott, D.J.; DeVos, S.; Pollaro, J.; Rooney, W.D.; Sweeney, H.L.; Bönnemann, C.G.; et al. Chemical shift-based MRI to measure fat fractions in dystrophic skeletal muscle. Magn. Reson. Med. 2014, 72, 8–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hausman, G.J.; Dodson, M.V.; Ajuwon, K.; Azain, M.; Barnes, K.M.; Guan, L.L.; Jiang, Z.; Poulos, S.P.; Sainz, R.D.; Smith, S.; et al. Board-invited review: The biology and regulation of preadipocytes and adipocytes in meat animals. J. Anim. Sci. 2009, 87, 1218–1246. [Google Scholar] [CrossRef] [Green Version]
- Ahn, B.; Soundarapandian, M.M.; Sessions, H.; Peddibhotla, S.; Roth, G.P.; Li, J.L.; Sugarman, E.; Koo, A.; Malany, S.; Wang, M.; et al. MondoA coordinately regulates skeletal myocyte lipid homeostasis and insulin signaling. J. Clin. Investig. 2016, 126, 3567–3579. [Google Scholar] [CrossRef]
- Kikuchi, K.; Fukuda, M.; Ito, T.; Inoue, M.; Yokoi, T.; Chiku, S.; Mitsuyama, T.; Asai, K.; Hirose, T.; Aizawa, Y. Transcripts of unknown function in multiple-signaling pathways involved in human stem cell differentiation. Nucleic Acids Res. 2009, 37, 4987–5000. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Pardo, G.; Sabater, M.; Hummel, M.; Ferrer, A.; Rodriguez-Hermosa, J.I.; Ruiz, B.; Ricart, W.; Peral, B.; et al. MiRNA Expression Profile of Human Subcutaneous Adipose and during Adipocyte Differentiation. PLoS ONE 2010, 5, e9022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Fang, L.; Liu, A.; Wang, X.; Zhao, B.; Wang, N. Activation of PPAR-δ induces microRNA-100 and decreases the uptake of very low-density lipoprotein in endothelial cells. Br. J. Pharmacol. 2015, 172, 3728–3736. [Google Scholar] [CrossRef] [PubMed]
- Estep, M.; Armistead, D.; Hossain, N.; Elarainy, H.; Goodman, Z.; Baranova, A.; Chandhoke, V.; Younossi, Z.M. Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 2010, 32, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Baranova, A.; Schlauch, K.; Elariny, H.; Jarrar, M.; Bennett, C.; Nugent, C.; Gowder, S.J.; Younoszai, Z.; Collantes, R.; Chandhoke, V.; et al. Gene expression patterns in hepatic tissue and visceral adipose tissue of patients with non-alcoholic fatty liver disease. Obes. Surg. 2007, 17, 1111–1118. [Google Scholar] [CrossRef]
- Eguchi, Y.; Mizuta, T.; Sumida, Y.; Ishibashi, E.; Kitajima, Y.; Isoda, H.; Horie, H.; Tashiro, T.; Iwamoto, E.; Takahashi, H.; et al. The pathological role of visceral fat accumulation in steatosis, inflammation, and progression of nonalcoholic fatty liver disease. J. Gastroenterol. 2011, 46, 70–78. [Google Scholar] [CrossRef]
- Smolka, C.; Schlösser, D.; Hohnloser, C.; Bemtgen, X.; Jänich, C.; Schneider, L.; Martin, J.; Pfeifer, D.; Moser, M.; Hasselblatt, P.; et al. miR-100 overexpression attenuates high fat diet induced weight gain, liver steatosis, hypertriglyceridemia and development of metabolic syndrome in mice. Mol. Med. 2021, 27, 101. [Google Scholar] [CrossRef] [PubMed]






| Oligonucleotide | Sequence |
|---|---|
| miR-100 WT-Sense | 5′pAAACTAGCGGCCGCTCCAGTAGATTACGGGTAGTCAGTTGAC TCTAG-3′ |
| miR-100 WT-Antisense | 3′-pTTTGATCGCCGGCGAGGTCATCTAATGCCCATCAGTCAACTG AGATC-5′ |
| miR-100 Mutant-Sense | 3′pTTTGATCGCCGGCGAGGTCATCTATTTTTTTTCAGTCAACTG AGATC-5′ |
| miR-100 Mutant-Antisense | 3′pTTTGATCGCCGGCGAGGTCATCTAAAAAAAAACAGTCAACTG AGATC-5′ |
| Gene | Accession Number | Forward Primer (5′) | Reverse Primer (3′) | Product Size, bp |
|---|---|---|---|---|
| IGF1R | NM_001244612.1 | CCAAAACCGAAGCTGAGAAG | TCCGGGTCTGTGATGTTGTA | 199 |
| MYOG | NM_001111325.1 | TGGGCGTGTAAGGTGTGTAA | TGCAGGCGCTCTATGTACTG | 197 |
| MYOD | NM_001040478.2 | TTTGCCAGAGCAGGAGCCCCTC | TTCGAACACCTGAGCGAGCGC | 123 |
| 16S | NC_006853.1 | CTTGTATGAATGGCCGCACG | GATGTAGCGGGTCGTAGTGG | 879 |
| HK2 | XM_015473383.2 | CTCAGAGCGGCTCAAGACAA | GCACACCTCCTTGACGATGA | 154 |
| COX7A2 | NM_175807.1 | ACTGAGCCAAGATGCTACGG | TGAAGCCACAGCCAGTTCAT | 242 |
| TFAM | NM_001034016.2 | GGCAGACTGGCAGGTATACAA | TGTGATGTGCCATCCCTAGC | 225 |
| CPT1B | NM_001034349.2 | CTCTCCACTAGCCAGATCGC | CGCTGGGCATTTGTCTCTGA | 197 |
| CD36 | NM_001278621.1 | GACGGATGTACAGCGGTGAT | GGTTGCCAAGAGGTCTGGTT | 250 |
| ACADM | NM_001075235.1 | CATGATCGCGCTGTTTAGGC | CAGGGTACTCGCCGGTTTTA | 224 |
| FABP31 | NM_174313 | GCGTTCTCTGTCGTCTTTCC | CTTGGTCATATTGCCCACCT | 154 |
| FABP41 | NM_174314.2 | GGATGGAAAATCAACCACCA | TGGACAACGTATCCAGCAGA | 174 |
| ELOVL6 | NM_001102155.1 | CATAGCACAGCCTCGGTCTA | TGCTGTGTCCTACCCCATTT | 172 |
| ACC2 | NM_001205333.2 | TCATTCACCTCCTGTCCACC | TAAACAGGAGTGAGCTGGGG | 196 |
| GAPDH | NM_001034034.2 | GGGTCATCTCTGCACCT | ACAGTCTTCTGGGTGGCAGT | 208 |
| Treatment | |||||||
|---|---|---|---|---|---|---|---|
| Negative Control | AgomiR | AntagomiR | p-Value | ||||
| Mean | SE | Mean | SE | Mean | SE | ||
| Fat particles/mm² | 1961.4 b | 196.0 | 2914.3 a | 163.0 | 2018.7 b | 30.2 | 0.007 |
| Fat particle distance, µm | 9.93 a | 0.37 | 8.89 b | 0.17 | 9.83 ab | 0.02 | 0.039 |
| Fat area percentage | 4.78 ab | 0.21 | 5.83 a | 0.54 | 3.99 b | 0.25 | 0.033 |
| Fat particle size, µm² | 24.45 | 1.55 | 19.71 | 0.65 | 19.57 | 1.16 | 0.043 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mir, B.A.; Albrecht, E.; Ali, A.; Hansson, O.; Maak, S. MicroRNA-100 Reduced Fetal Bovine Muscle Satellite Cell Myogenesis and Augmented Intramuscular Lipid Deposition by Modulating IGF1R. Cells 2022, 11, 451. https://doi.org/10.3390/cells11030451
Mir BA, Albrecht E, Ali A, Hansson O, Maak S. MicroRNA-100 Reduced Fetal Bovine Muscle Satellite Cell Myogenesis and Augmented Intramuscular Lipid Deposition by Modulating IGF1R. Cells. 2022; 11(3):451. https://doi.org/10.3390/cells11030451
Chicago/Turabian StyleMir, Bilal Ahmad, Elke Albrecht, Asghar Ali, Ola Hansson, and Steffen Maak. 2022. "MicroRNA-100 Reduced Fetal Bovine Muscle Satellite Cell Myogenesis and Augmented Intramuscular Lipid Deposition by Modulating IGF1R" Cells 11, no. 3: 451. https://doi.org/10.3390/cells11030451
APA StyleMir, B. A., Albrecht, E., Ali, A., Hansson, O., & Maak, S. (2022). MicroRNA-100 Reduced Fetal Bovine Muscle Satellite Cell Myogenesis and Augmented Intramuscular Lipid Deposition by Modulating IGF1R. Cells, 11(3), 451. https://doi.org/10.3390/cells11030451