Functional Genomic Screening in Human Pluripotent Stem Cells Reveals New Roadblocks in Early Pancreatic Endoderm Formation
Abstract
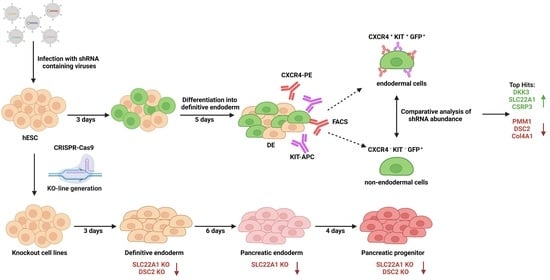
:1. Introduction
2. Materials and Methods
2.1. shRNA Screen
2.2. Stem Cell Culture
2.3. Generation of Knockout Cell Lines
2.4. DNA Isolation, PCR and Sequencing
2.5. Differentiation into Pancreatic Progenitor Cells
2.6. RNA Isolation and qPCR
2.7. Flow Cytometry
2.8. ICC Staining
2.9. Bioinformatics Analysis
2.10. Statistical Analysis
3. Results
3.1. RNA Interference Approach during Endodermal Differentiation of Human PSCs
3.2. Newly Generated Knockout Cell Lines Still Express Pluripotency Markers
3.3. Knockout of DSC2 and SLC22A1 Leads to Impaired DE Formation
3.4. Pancreatic Progenitor Formation Is Affected by DSC2 and SLC22A1 Loss
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bastidas-Ponce, A.; Scheibner, K.; Lickert, H.; Bakhti, M. Cellular and molecular mechanisms coordinating pancreas development. Development 2017, 144, 2873–2888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiecker, C.; Bates, T.; Bell, E. Molecular specification of germ layers in vertebrate embryos. Cell Mol. Life Sci. 2016, 73, 923–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okabe, M. The Cell Biology of Mammalian Fertilization. Development 2013, 140, 4471–4479. [Google Scholar] [CrossRef] [Green Version]
- Wamaitha, S.E.; Niakan, K.K. Human Pre-Gastrulation Development. Curr. Top Dev. Biol. 2018, 128, 295–338. [Google Scholar] [PubMed]
- Zorn, A.M.; Wells, J.M. Vertebrate Endoderm Development and Organ Formation. Annu. Rev. Cell Dev. Biol. 2009, 25, 221–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiyaboonchai, A.; Cardenas-Diaz, F.L.; Ying, L.; Maguire, J.A.; Sim, X.; Jobaliya, C.; Gagne, A.L.; Kishore, S.; Stanescu, D.E.; Hughes, N.; et al. Gata6 Plays an Important Role in the Induction of Human Definitive Endoderm, Development of the Pancreas, and Functionality of Pancreatic Β cells. Stem Cell Rep. 2017, 8, 589–604. [Google Scholar] [CrossRef]
- Zhu, Z.; Huangfu, D. Human Pluripotent Stem Cells: An Emerging Model in Developmental Biology. Development 2013, 140, 705–717. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.-D.; Lee, K.; Yang, D.; Amin, S.; Verma, N.; Li, Q.V.; Zhu, Z.; Soh, C.-L.; Kumar, R.; Evans, T.; et al. Genome Editing in Hpscs Reveals Gata6 Haploinsufficiency and a Genetic Interaction with Gata4 in Human Pancreatic Development. Cell Stem Cell 2017, 20, 675–688.e6. [Google Scholar] [CrossRef] [Green Version]
- Jennings, R.E.; Berry, A.A.; Kirkwood-Wilson, R.; Roberts, N.A.; Hearn, T.; Salisbury, R.J.; Blaylock, J.; Hanley, K.P.; Hanley, N.A. Development of the Human Pancreas from Foregut to Endocrine Commitment. Diabetes 2013, 62, 3514–3522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennings, R.E.; Berry, A.A.; Strutt, J.P.; Gerrard, D.; Hanley, N.A. Human pancreas development. Development 2015, 142, 3126–3137. [Google Scholar] [CrossRef] [Green Version]
- Ramond, C.; Beydag-Tasöz, B.S.; Azad, A.; van de Bunt, M.; Petersen, M.B.K.; Beer, N.L.; Glaser, N.; Berthault, C.; Gloyn, A.L.; Hansson, M.; et al. Understanding Human Fetal Pancreas Development Using Subpopulation Sorting, Rna Sequencing and Single-Cell Profiling. Development 2018, 145, dev165480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yiangou, L.; Ross, A.D.; Goh, K.J.; Vallier, L. Human Pluripotent Stem Cell-Derived Endoderm for Modeling Development and Clinical Applications. Cell Stem Cell 2018, 22, 485–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed Differentiation of Human Pluripotent Stem Cells into Intestinal Tissue In Vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hohwieler, M.; Renz, S.; Liebau, S.; Lin, Q.; Lechel, A.; Klaus, J.; Perkhofer, L.; Zenke, M.; Seufferlein, T.; Illing, A.; et al. “Miniguts” from Plucked Human Hair Meet Crohn’s Disease. Z. Gastroenterol. 2016, 54, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Krüger, J.; Groß, R.; Conzelmann, V.; Müller, J.A.; Koepke, L.; Sparrer, K.M.J.; Weil, T.; Schütz, D.; Seufferlein, T.; Barth, T.F.E.; et al. Drug Inhibition of Sars-Cov-2 Replication in Human Pluripotent Stem Cell-Derived Intestinal Organoids. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 935–948. [Google Scholar] [CrossRef]
- Mun, S.J.; Ryu, J.-S.; Lee, M.-O.; Son, Y.S.; Oh, S.J.; Cho, H.-S.; Son, M.-Y.; Kim, D.-S.; Kim, S.J.; Yoo, H.J.; et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019, 71, 970–985. [Google Scholar] [CrossRef]
- Breunig, M.; Merkle, J.; Wagner, M.; Melzer, M.K.; Barth, T.F.E.; Engleitner, T.; Krumm, J.; Wiedenmann, S.; Cohrs, C.M.; Perkhofer, L.; et al. Modeling plasticity and dysplasia of pancreatic ductal organoids derived from human pluripotent stem cells. Cell Stem Cell 2021, 28, 1105–1124.e19. [Google Scholar] [CrossRef]
- Rezania, A.; Bruin, J.; Arora, P.; Rubin, A.; Batushansky, I.; Asadi, A.; O’Dwyer, S.; Quiskamp, N.; Mojibian, M.; Albrecht, T.; et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014, 32, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Hohwieler, M.; Illing, A.; Hermann, P.C.; Mayer, T.; Stockmann, M.; Perkhofer, L.; Eiseler, T.; Antony, J.S.; Müller, M.; Renz, S.; et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 2017, 66, 473–486. [Google Scholar] [CrossRef] [Green Version]
- Millman, J.R.; Xie, C.; Van Dervort, A.; Gürtler, M.; Pagliuca, F.W.; Melton, D.A. Melton. Generation of stem cell-derived Β-cells from patients with type 1 diabetes. Nat. Commun. 2016, 7, 11463. [Google Scholar] [CrossRef] [Green Version]
- Hogrebe, N.J.; Augsornworawat, P.; Maxwell, K.G.; Velazco-Cruz, L.; Millman, J.R. Targeting the cytoskeleton to direct pancreatic differentiation of human pluripotent stem cells. Nat. Biotechnol. 2020, 38, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Veres, A.; Faust, A.L.; Bushnell, H.; Engquist, E.; Kenty, J.H.-R.; Harb, G.; Poh, Y.-C.; Sintov, E.; Gürtler, M.; Pagliuca, F.W.; et al. Charting Cellular Identity During Human in Vitro Β-Cell Differentiation. Nature 2019, 569, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Naxerova, K.; Di Stefano, B.; Makofske, J.L.; Watson, E.V.; de Kort, M.A.; Martin, T.D.; Dezfulian, M.; Ricken, D.; Wooten, E.C.; Kuroda, M.I.; et al. Integrated loss- and gain-of-function screens define a core network governing human embryonic stem cell behavior. Genes Dev. 2021, 35, 1527–1547. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.; Mahaddalkar, P.U.; Kraus, J.M.; Zhong, X.; Bergmann, W.; Srinivasan, D.; Gout, J.; Roger, E.; Beutel, A.K.; Zizer, E.; et al. Functional genomic screening during somatic cell reprogramming identifies Dkk3 as a roadblock of organ regeneration. Adv. Sci. 2021, 8, 2100626. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.; Li, M.Z.; Chang, K.; Ge, W.; Golding, M.; Rickles, R.J.; Siolas, D.; Hu, G.; Paddison, P.J.; Schlabach, M.R.; et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005, 37, 1281–1288. [Google Scholar] [CrossRef]
- Nostro, M.C.; Sarangi, F.; Yang, C.; Holland, A.; Elefanty, A.G.; Stanley, E.G.; Greiner, D.L.; Keller, G. Efficient Generation of NKX6-1+ Pancreatic Progenitors from Multiple Human Pluripotent Stem Cell Lines. Stem Cell Rep. 2015, 4, 591–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breunig, M.; Merkle, J.; Melzer, M.K.; Heller, S.; Seufferlein, T.; Meier, M.; Hohwieler, M.; Kleger, A. Differentiation of human pluripotent stem cells into pancreatic duct-like organoids. STAR Protoc. 2021, 2, 100913. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for Rna-Seq data with Deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Philippi, A.; Heller, S.; Costa, I.G.; Senée, V.; Breunig, M.; Li, Z.; Kwon, G.; Russell, R.; Illing, A.; Lin, Q.; et al. Mutations and variants of ONECUT1 in diabetes. Nat. Med. 2021, 27, 1928–1940. [Google Scholar] [CrossRef]
- Heller, S.; Li, Z.; Lin, Q.; Geusz, R.; Breunig, M.; Hohwieler, M.; Zhang, X.; Nair, G.G.; Seufferlein, T.; Hebrok, M.; et al. Transcriptional Changes and the Role of Onecut1 in Hpsc Pancreatic differentiation. Commun. Biol. 2021, 4, 1298. [Google Scholar] [CrossRef]
- Miyanari, Y.; Torres-Padilla, M.-E. Control of Ground-State Pluripotency by Allelic Regulation of Nanog. Nature 2012, 483, 470–473. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Wiedenmann, S.; Breunig, M.; Merkle, J.; von Toerne, C.; Georgiev, T.; Moussus, M.; Schulte, L.; Seufferlein, T.; Sterr, M.; Lickert, H.; et al. Single-cell-resolved differentiation of human induced pluripotent stem cells into pancreatic duct-like organoids on a microwell chip. Nat. Biomed. Eng. 2021, 5, 897–913. [Google Scholar] [CrossRef]
- D’Amour, K.A.; Agulnick, A.D.; Eliazer, S.; Kelly, O.G.; Kroon, E.; E Baetge, E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005, 23, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Mahaddalkar, P.U.; Scheibner, K.; Pfluger, S.; Ansarullah; Sterr, M.; Beckenbauer, J.; Irmler, M.; Beckers, J.; Knöbel, S.; Lickert, H. Generation of Pancreatic Β Cells from Cd177(+) Anterior Definitive Endoderm. Nat. Biotechnol. 2020, 38, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Teo, A.K.K.; Ali, Y.; Wong, K.Y.; Chipperfield, H.; Sadasivam, A.; Poobalan, Y.; Tan, E.K.; Wang, S.T.; Abraham, S.; Tsuneyoshi, N.; et al. Activin and Bmp4 synergistically promote formation of definitive endoderm in human embryonic stem cells. Stem Cells 2012, 30, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Burtscher, I.; Lickert, H. Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development 2009, 136, 1029–1038. [Google Scholar] [CrossRef] [Green Version]
- Liew, L.C.; Gailhouste, L.; Tan, G.C.; Yamamoto, Y.; Takeshita, F.; Nakagama, H.; Ochiya, T. Microrna-124a Inhibits Endoderm Lineage Commitment by Targeting Sox17 and Gata6 in Mouse Embryonic Stem Cells. Stem Cells 2020, 38, 504–515. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Lu, P.; Li, M.; Yan, C.; Zhang, T.; Jiang, W. Gata6-As1 Regulates Gata6 Expression to Modulate Human Endoderm Differentiation. Stem Cell Rep. 2020, 15, 694–705. [Google Scholar] [CrossRef]
- Delva, E.; Tucker, D.K.; Kowalczyk, A.P. The Desmosome. Cold Spring Harb. Perspect. Biol. 2009, 1, a002543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuser, A.; Plovie, E.R.; Ellinor, P.; Grossmann, K.S.; Shin, J.T.; Wichter, T.; Basson, C.T.; Lerman, B.B.; Sasse-Klaassen, S.; Thierfelder, L.; et al. Mutant Desmocollin-2 causes Arrhythmogenic right ventricular cardiomyopathy. Am. J. Hum. Genet. 2006, 79, 1081–1088. [Google Scholar] [CrossRef] [Green Version]
- Syrris, P.; Ward, D.; Evans, A.; Asimaki, A.; Gandjbakhch, E.; Sen-Chowdhry, S.; McKenna, W.J. Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Associated with Mutations in the Desmosomal Gene Desmocollin-2. Am. J. Hum. Genet. 2006, 79, 978–984. [Google Scholar] [CrossRef] [Green Version]
- Brodehl, A.; Weiss, J.; Debus, J.D.; Stanasiuk, C.; Klauke, B.; Deutsch, M.A.; Fox, H.; Bax, J.; Ebbinghaus, H.; Gärtner, A.; et al. A homozygous DSC2 deletion associated with arrhythmogenic cardiomyopathy is caused by uniparental isodisomy. J. Mol. Cell. Cardiol. 2020, 141, 17–29. [Google Scholar] [CrossRef]
- Fang, W.-K.; Liao, L.-D.; Zeng, F.-M.; Zhang, P.-X.; Wu, J.-Y.; Shen, J.; Xu, L.-Y.; Li, E.-M. Desmocollin-2 affects the adhesive strength and cytoskeletal arrangement in esophageal squamous cell carcinoma cells. Mol. Med. Rep. 2014, 10, 2358–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, W.-K.; Gu, W.; Li, E.-M.; Wu, Z.-Y.; Shen, Z.-Y.; Shen, J.-H.; Wu, J.-Y.; Pan, F.; Lv, Z.; Xu, X.-E.; et al. Reduced membranous and ectopic cytoplasmic expression of DSC2 in esophageal squamous cell carcinoma: An independent prognostic factor. Hum. Pathol. 2010, 41, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.; Mansour, S.; Ahnood, D.; Kalidas, K.; Patton, M.; McKenna, W.J.; Behr, E.; Crosby, A. Homozygous mutation of Desmocollin-2 in Arrhythmogenic right ventricular cardiomyopathy with mild palmoplantar keratoderma and woolly hair. Cardiology 2009, 113, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.; Hatzfeld, M.; Keil, R. Desmosomes as signaling hubs in the regulation of cell behavior. Front. Cell Dev. Biol. 2021, 9, 745670. [Google Scholar] [CrossRef] [PubMed]
- De Franco, E.; Shaw-Smith, C.; Flanagan, S.E.; Shepherd, M.H.; Hattersley, A.T.; Ellard, S. Gata6 Mutations Cause a Broad Phenotypic Spectrum of Diabetes from Pancreatic Agenesis to Adult-Onset Diabetes without Exocrine Insufficiency. Diabetes 2013, 62, 993–997. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.T.; Moore, L.; Poplawski, N.K.; De Sousa, S.M.C. Familial Gata6 Mutation Causing Variably Expressed Diabetes Mellitus and Cardiac and Renal Abnormalities. Endocrinol. Diabetes Metab. Case Rep. 2019, 2019. [Google Scholar] [CrossRef]
- Villamayor, L.; Rodríguez-Seguel, E.; Araujo, R.; Carrasco, M.; Bru-Tarí, E.; Mellado-Gil, J.M.; Gauthier, B.R.; Martinelli, P.; Quesada, I.; Soria, B.; et al. Gata6 controls Insulin Biosynthesis and secretion in adult Β-Cells. Diabetes 2018, 67, 448–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyrsova, L.; Smutný, T.; Trejtnar, F.; Pavek, P. Expression of organic cation Transporter 1 (Oct1): Unique patterns of indirect regulation by nuclear receptors and hepatospecific gene regulation. Drug Metab. Rev. 2016, 48, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Herraez, E.; Lozano, E.; Macias, R.I.; Vaquero, J.; Bujanda, L.; Banales, J.M.; Marin, J.J.; Briz, O. Expression ofSLC22A1variants may affect the response of hepatocellular carcinoma and cholangiocarcinoma to sorafenib. Hepatology 2013, 58, 1065–1073. [Google Scholar] [CrossRef]
- Gromicho, M.; Dinis, J.; Magalhães, M.; Fernandes, A.R.; Tavares, P.; Laires, A.; Rueff, J.; Rodrigues, A.S. Development of Imatinib and Dasatinib Resistance: Dynamics of Expression of Drug Transporters Abcb1, Abcc1, Abcg2, Mvp, and Slc22a1. Leuk. Lymphoma 2011, 52, 1980–1990. [Google Scholar] [CrossRef]
- Dawed, A.Y.; Zhou, K.; van Leeuwen, N.; Mahajan, A.; Robertson, N.; Koivula, R.; Elders, P.J.; Rauh, S.P.; Jones, A.G.; Holl, R.W.; et al. Variation in the plasma membrane monoamine transporter (Pmat) (Encoded by Slc29a4) and organic cation transporter 1 (Oct1) (Encoded by Slc22a1) and gastrointestinal intolerance to metformin in type 2 diabetes: An Imi direct study. Diabetes Care 2019, 42, 1027–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dujic, T.; Zhou, K.; Donnelly, L.A.; Tavendale, R.; Palmer, C.N.; Pearson, E.R. Association of Organic Cation Transporter 1 with Intolerance to Metformin in Type 2 Diabetes: A Godarts Study. Diabetes 2015, 64, 1786–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Gene | Clone | Allele 1 | Allele 2 |
|---|---|---|---|
| DKK3 | 1 | +1 bp insertion | +1 bp insertion |
| 2 | +1 bp insertion | −2 bp deletion | |
| SLC22A1 | 1 | −1 bp deletion | −8 bp deletion |
| 2 | +1 bp insertion | −1 bp deletion | |
| CSRP3 | 1 | +1 pb insertion | −4 bp deletion, +10 bp insertion |
| 2 | −1 bp deletion | −26 bp deletion | |
| PMM1 | 1 | +1 bp insertion | +1 bp insertion |
| 2 | −7 bp deletion | −8 bp deletion | |
| DSC2 | 1 | −2 bp deletion | −11 bp deletion |
| 2 | −1 bp deletion | −10 bp deletion | |
| COL4A1 | 1 | −1 bp deletion | +1 bp insertion |
| 2 | +1 bp insertion | +1 bp insertion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krüger, J.; Breunig, M.; Pasquini, L.P.; Morawe, M.; Groß, A.; Arnold, F.; Russell, R.; Seufferlein, T.; Azoitei, N.; Kestler, H.A.; et al. Functional Genomic Screening in Human Pluripotent Stem Cells Reveals New Roadblocks in Early Pancreatic Endoderm Formation. Cells 2022, 11, 582. https://doi.org/10.3390/cells11030582
Krüger J, Breunig M, Pasquini LP, Morawe M, Groß A, Arnold F, Russell R, Seufferlein T, Azoitei N, Kestler HA, et al. Functional Genomic Screening in Human Pluripotent Stem Cells Reveals New Roadblocks in Early Pancreatic Endoderm Formation. Cells. 2022; 11(3):582. https://doi.org/10.3390/cells11030582
Chicago/Turabian StyleKrüger, Jana, Markus Breunig, Lino Pascal Pasquini, Mareen Morawe, Alexander Groß, Frank Arnold, Ronan Russell, Thomas Seufferlein, Ninel Azoitei, Hans A. Kestler, and et al. 2022. "Functional Genomic Screening in Human Pluripotent Stem Cells Reveals New Roadblocks in Early Pancreatic Endoderm Formation" Cells 11, no. 3: 582. https://doi.org/10.3390/cells11030582
APA StyleKrüger, J., Breunig, M., Pasquini, L. P., Morawe, M., Groß, A., Arnold, F., Russell, R., Seufferlein, T., Azoitei, N., Kestler, H. A., Julier, C., Heller, S., Hohwieler, M., & Kleger, A. (2022). Functional Genomic Screening in Human Pluripotent Stem Cells Reveals New Roadblocks in Early Pancreatic Endoderm Formation. Cells, 11(3), 582. https://doi.org/10.3390/cells11030582