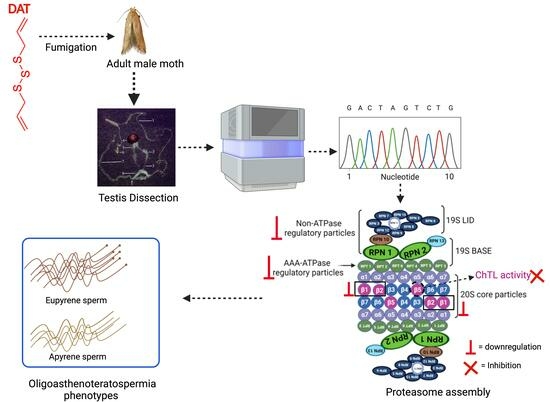
Diallyl Trisulfide Causes Male Infertility with Oligoasthenoteratospermia in Sitotroga cerealella through the Ubiquitin–Proteasome Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Test Animals and DAT Fumigation
2.2. Light Microscopy of Testes and Spermatophore
2.3. Morphological Analysis of Spermatophore Formation
2.4. RNA Extraction, cDNA Library Preparation, and Sequencing
2.5. Transcriptome Assembly and Annotation
2.6. DEG Detection and Functional Enrichment Analysis
2.7. Quantitative Real-Time PCR (qRT-PCR)
2.8. Western Blotting
2.9. Determination of 26S Proteasome Activation Assay
2.10. Detection of Aggresomes
2.11. Statistical Analysis
3. Results
3.1. DAT Caused Disruption in Testis and Sperm Bundle Morphology of S. cerealella
3.2. DAT Induces Abnormal Spermatophore Formation in S. cerealella
3.3. RNA-Seq Revealed the Global Downregulation of Gene Expression in Response to DAT Fumigation
3.4. GO and KEGG Downregulated Genes Enriched in the Ubiquitin Proteasome System (UPS)
3.5. Validation of RNA-Seq Data and the Effects of DAT on the Expression of Proteasome Subunits
3.6. DAT Inhibited the Expression of the Proteasome Catalytic Core Complex
3.7. DAT Inhibited 26S Proteasome Activity and Formed Aggresomal Proteins in the Testes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Athanassiou, C.G.; Rumbos, C.I. Emerging pests in durable stored products. In Recent Advances in Stored Product Protection; Springer: Berlin/Heidelberg, Germany, 2018; pp. 211–227. [Google Scholar]
- Bushra, S.; Aslam, M. Management of Sitotroga cerealella in stored cereal grains: A review. Arch. Phytopathol. Plant Prot. 2014, 47, 2365–2376. [Google Scholar] [CrossRef]
- Chang, M.M.; Shah, S.; Wu, M.Y.; Zhang, S.S.; Wu, G.; Yang, F.L. Effect of diallyl trisulfide on the reproductive behavior of the grain moth, Sitotroga cerealella (Lepidoptera: Gelechiidae). Insects 2020, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Nayak, M.K.; Daglish, G.J.; Phillips, T.W.; Ebert, P.R. Resistance to the fumigant phosphine and its management in insect pests of stored products: A global perspective. Annu. Rev. Entomol. 2020, 65, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Li, S.; Wu, H.; Gao, Q.; Shi, S.; Huang, Y.; Cao, H. Transcriptomic analysis of Sitophilus zeamais in response to limonene fumigation. Pest Manag. Sci. 2022, 78, 4774–4782. [Google Scholar] [CrossRef]
- Yang, F.-L.; Li, X.-G.; Zhu, F.; Lei, C.-L. Structural Characterization of Nanoparticles Loaded with Garlic Essential Oil and Their Insecticidal Activity against Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae). J. Agric. Food Chem. 2009, 57, 10156–10162. [Google Scholar] [CrossRef]
- Malla, R.; Marni, R.; Chakraborty, A.; Kamal, M.A. Diallyl disulfide and diallyl trisulfide in garlic as novel therapeutic agents to overcome drug resistance in breast cancer. J. Pharm. Anal. 2021, 12, 221–231. [Google Scholar] [CrossRef]
- Jeremic, J.N.; Jakovljevic, V.L.; Zivkovic, V.I.; Srejovic, I.M.; Bradic, J.V.; Milosavljevic, I.M.; Mitrovic, S.L.; Jovicic, N.U.; Bolevich, S.B.; Svistunov, A.A. Garlic derived diallyl trisulfide in experimental metabolic syndrome: Metabolic effects and cardioprotective role. Int. J. Mol. Sci. 2020, 21, 9100. [Google Scholar] [CrossRef]
- Zhu, X.; Lu, R.; Zhang, G.; Fan, L.; Zhan, Y.; Chen, G.; Zhou, L. Diallyl Trisulfide attenuates alcohol-induced hepatocyte pyroptosis via elevation of hydrogen sulfide. Biosci. Biotechnol. Biochem. 2022, 86, 1552–1561. [Google Scholar] [CrossRef]
- Shah, S.; Hafeez, M.; Wu, M.-Y.; Zhang, S.-S.; Ilyas, M.; Wu, G.; Yang, F.-L. Downregulation of chitin synthase A gene by diallyl trisulfide, an active substance from garlic essential oil, inhibits oviposition and alters the morphology of adult Sitotroga cerealella. J. Pest Sci. 2020, 93, 1097–1106. [Google Scholar] [CrossRef]
- Shah, S.; Zhang, S.-S.; Elgizawy, K.K.; Yan, W.-H.; Tang, N.; Wu, G.; Yang, F.-L. Diallyl trisulfide reduced the reproductive capacity of male Sitotroga cerealella via the regulation of juvenile and ecdysone hormones. Ecotoxicol. Environ. Saf. 2022, 248, 114304. [Google Scholar] [CrossRef]
- Shah, S.; Elgizawy, K.K.; Shi, C.-M.; Yao, H.; Yan, W.-H.; Li, Y.; Wang, X.-P.; Wu, G.; Yang, F.-L. Diallyl Trisulfide, a Biologically Active Component of Garlic Essential Oil, Decreases Male Fertility in Sitotroga cerealella by Impairing Dimorphic Spermatogenesis, Sperm Motility and Lipid Homeostasis. Cells 2023, 12, 669. [Google Scholar] [CrossRef] [PubMed]
- Glotzer, M.; Murray, A.W.; Kirschner, M.W. Cyclin is degraded by the ubiquitin pathway. Nature 1991, 349, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Lander, G.C.; Estrin, E.; Matyskiela, M.E.; Bashore, C.; Nogales, E.; Martin, A. Complete subunit architecture of the proteasome regulatory particle. Nature 2012, 482, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Seemuller, E.; Lupas, A.; Stock, D.; Lowe, J.; Huber, R.; Baumeister, W. Proteasome from Thermoplasma acidophilum: A threonine protease. Science 1995, 268, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Pathare, G.R.; Nagy, I.; Śledź, P.; Anderson, D.J.; Zhou, H.-J.; Pardon, E.; Steyaert, J.; Förster, F.; Bracher, A.; Baumeister, W. Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc. Natl. Acad. Sci. USA 2014, 111, 2984–2989. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Lee, B.-H.; Hanna, J.; King, R.W.; Finley, D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol. Cell. Proteom. 2011, 10, R110.003871. [Google Scholar] [CrossRef]
- Tomko, R.J., Jr.; Hochstrasser, M. Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 2013, 82, 415–445. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Sutovsky, P. Sperm proteasome and fertilization. Reproduction 2011, 142, 1. [Google Scholar] [CrossRef]
- Bose, R.; Manku, G.; Culty, M.; Wing, S.S. Ubiquitin–proteasome system in spermatogenesis. Posttransl. Protein Modif. Reprod. Syst. 2014, 759, 181–213. [Google Scholar]
- Culling, C.F.A. Handbook of Histopathological and Histochemical Techniques: Including Museum Techniques; Butterworth-Heinemann: Oxford, UK, 2013; ISBN 1483164799. [Google Scholar]
- McFadden, T.; Devulapalli, R.K.; Jarome, T.J. Quantifying subcellular ubiquitin-proteasome activity in the rodent brain. JoVE J. Vis. Exp. 2019, e59695. [Google Scholar]
- Coelho, D.S.; Schwartz, S.; Merino, M.M.; Hauert, B.; Topfel, B.; Tieche, C.; Rhiner, C.; Moreno, E. Culling Less Fit Neurons Protects against Amyloid-β-Induced Brain Damage and Cognitive and Motor Decline. Cell Rep. 2018, 25, 3661–3673.e3. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Li, H.-M.; Yang, W.-J.; Wei, D.-D.; Dou, W.; Huang, Y.; Wang, J.-J. Transcriptome Profiling of the Testis Reveals Genes Involved in Spermatogenesis and Marker Discovery in the Oriental Fruit Fly, Bactrocera Dorsalis. Insect Mol. Biol. 2015, 24, 41–57. [Google Scholar] [CrossRef]
- Vedelek, V.; Bodai, L.; Grézal, G.; Kovács, B.; Boros, I.M.; Laurinyecz, B.; Sinka, R. Analysis of Drosophila Melanogaster Testis Transcriptome. BMC Genom. 2018, 19, 697. [Google Scholar] [CrossRef]
- Zhong, L.; Belote, J.M. The Testis-Specific Proteasome Subunit Prosα6T of D. Melanogaster Is Required for Individualization and Nuclear Maturation during Spermatogenesis. Development 2007, 134, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, R.-R.; Peng, L.-Y.; Zhang, L.; Yao, Y.-L.; Bao, Y.-Y. Proteolytic Activity of the Proteasome Is Required for Female Insect Reproduction. Open Biol. 2021, 11, 200251. [Google Scholar] [CrossRef] [PubMed]
- Whittington, E.; Zhao, Q.; Borziak, K.; Walters, J.R.; Dorus, S. Characterisation of the Manduca Sexta Sperm Proteome: Genetic Novelty Underlying Sperm Composition in Lepidoptera. Insect Biochem. Mol. Biol. 2015, 62, 183–193. [Google Scholar] [CrossRef]
- Whittington, E.; Forsythe, D.; Borziak, K.; Karr, T.L.; Walters, J.R.; Dorus, S. Contrasting Patterns of Evolutionary Constraint and Novelty Revealed by Comparative Sperm Proteomic Analysis in Lepidoptera. BMC Genom. 2017, 18, 931. [Google Scholar] [CrossRef]
- Chapman, R.F. The Insects: Structure and Function; Cambridge University Press: Cambridge, UK, 1998; ISBN 0521578906. [Google Scholar]
- Yan, W.; Wu, M.-Y.; Shah, S.; Yao, Y.-C.; Elgizawy, K.K.; Tang, N.; Wu, G.; Yang, F.-L. Silencing the triacylglycerol lipase (TGL) gene decreases the number of apyrene sperm and inhibits oviposition in Sitotroga cerealella. Cell. Mol. Life Sci. 2022, 79, s00018–s00021. [Google Scholar] [CrossRef]
- Wu, M.-Y.; Ying, Y.-Y.; Zhang, S.-S.; Li, X.-G.; Yan, W.-H.; Yao, Y.-C.; Shah, S.; Wu, G.; Yang, F.-L. Effects of diallyl trisulfide, an active substance from garlic essential oil, on energy metabolism in male moth Sitotroga cerealella olivier. Insects 2020, 11, 270. [Google Scholar] [CrossRef]
- Werner, M.; Tscheulin, T.; Speck, T.; Zissler, D.; Peschke, K. Ultrastructure and motility pattern of the spermatozoa of Aleochara curtula (Coleoptera, Staphylinidae). Arthropod Struct. Dev. 2002, 31, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Sonenshine, D.E.; Bissinger, B.W.; Egekwu, N.; Donohue, K.V.; Khalil, S.M.; Roe, R.M. First transcriptome of the testis-vas deferens-male accessory gland and proteome of the spermatophore from Dermacentor variabilis (Acari: Ixodidae). PLoS ONE 2011, 6, e24711. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.J.; Burns, A.C.; Kazi, A.; Ping Dou, Q. Direct Inhibition of the Ubiquitin–Proteasome Pathway by Ester Bond-Containing Green Tea Polyphenols Is Associated with Increased Expression of Sterol Regulatory Element-Binding Protein 2 and LDL Receptor. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2004, 1682, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Sun, J.; Xu, Q.; Liu, Y.; Wei, J.; Young, C.Y.F.; Yuan, H.; Lou, H. Marchantin M: A novel inhibitor of proteasome induces autophagic cell death in prostate cancer cells. Cell Death Dis. 2013, 4, e761. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Monsarrat, B.; Gairin, J.E.; Girbal-Neuhauser, E. Effect of ajoene, a natural antitumor small molecule, on human 20S proteasome activity in vitro and in human leukemic HL60 cells. Fundam. Clin. Pharmacol. 2004, 18, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wei, X.; Liu, L.; Sun, H.; Fang, T.; Wang, L.; Li, Y.; Sui, W.; Wang, K.; He, Y.; et al. Indirubin-3′-monoxime acts as proteasome inhibitor: Therapeutic application in multiple myeloma. eBioMedicine 2022, 78, 103950. [Google Scholar] [CrossRef]
- Zhang, Q.; Ji, S.-Y.; Busayavalasa, K.; Shao, J.; Yu, C. Meiosis I progression in spermatogenesis requires a type of testis-specific 20S core proteasome. Nat. Commun. 2019, 10, 3387. [Google Scholar] [CrossRef]
- Dickins, R.A.; Frew, I.J.; House, C.M.; O’Bryan, M.K.; Holloway, A.J.; Haviv, I.; Traficante, N.; de Kretser, D.M.; Bowtell, D.D.L. The ubiquitin ligase component Siah1a is required for completion of meiosis I in male mice. Mol. Cell. Biol. 2002, 22, 2294–2303. [Google Scholar] [CrossRef]
- Kerns, K.; Morales, P.; Sutovsky, P. Regulation of sperm capacitation by the 26S proteasome: An emerging new paradigm in spermatology. Biol. Reprod. 2016, 94, 111–117. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, S.; Elgizawy, K.K.; Wu, M.-Y.; Yao, H.; Yan, W.-H.; Li, Y.; Wang, X.-P.; Wu, G.; Yang, F.-L. Diallyl Trisulfide Causes Male Infertility with Oligoasthenoteratospermia in Sitotroga cerealella through the Ubiquitin–Proteasome Pathway. Cells 2023, 12, 2507. https://doi.org/10.3390/cells12202507
Shah S, Elgizawy KK, Wu M-Y, Yao H, Yan W-H, Li Y, Wang X-P, Wu G, Yang F-L. Diallyl Trisulfide Causes Male Infertility with Oligoasthenoteratospermia in Sitotroga cerealella through the Ubiquitin–Proteasome Pathway. Cells. 2023; 12(20):2507. https://doi.org/10.3390/cells12202507
Chicago/Turabian StyleShah, Sakhawat, Karam Khamis Elgizawy, Meng-Ya Wu, Hucheng Yao, Wen-Han Yan, Yu Li, Xiao-Ping Wang, Gang Wu, and Feng-Lian Yang. 2023. "Diallyl Trisulfide Causes Male Infertility with Oligoasthenoteratospermia in Sitotroga cerealella through the Ubiquitin–Proteasome Pathway" Cells 12, no. 20: 2507. https://doi.org/10.3390/cells12202507
APA StyleShah, S., Elgizawy, K. K., Wu, M. -Y., Yao, H., Yan, W. -H., Li, Y., Wang, X. -P., Wu, G., & Yang, F. -L. (2023). Diallyl Trisulfide Causes Male Infertility with Oligoasthenoteratospermia in Sitotroga cerealella through the Ubiquitin–Proteasome Pathway. Cells, 12(20), 2507. https://doi.org/10.3390/cells12202507