Spatial Transcriptomics Reveals Signatures of Histopathological Changes in Muscular Sarcoidosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Samples
2.2. Standard Protocol Approvals, Registrations and Patients Consents
2.3. Antibodies
2.4. Histology and Immunohistology
2.5. Spatial Transcriptomics Using Visium
2.6. Statistical Analysis and Software
3. Results
3.1. Cases Selection
3.2. Spatial Transcriptomics Analysis Identifies Five Different Clusters in Muscle Tissue Sections
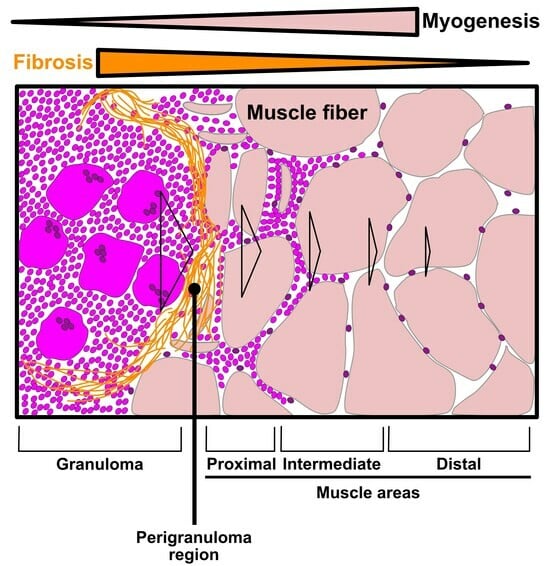
3.3. Evaluation of the Granuloma Structure’s Impact on the Surrounding Muscle Tissue through a Spatial Transcriptomics Approach
3.4. The Granuloma Structure Generates a Gradient of Transcriptomic Alterations in Muscle Fibers
3.5. The Granuloma Structure Is Characterized by a Pro-Fibrotic Macrophage Signature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sève, P.; Pacheco, Y.; Durupt, F.; Jamilloux, Y.; Gerfaud-Valentin, M.; Isaac, S.; Boussel, L.; Calender, A.; Androdias, G.; Valeyre, D.; et al. Sarcoidosis: A Clinical Overview from Symptoms to Diagnosis. Cells 2021, 10, 766. [Google Scholar] [CrossRef]
- Zhang, H.; Costabel, U.; Dai, H. The Role of Diverse Immune Cells in Sarcoidosis. Front. Immunol. 2021, 12, 788502. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.S.; Moller, D.R. Etiology of Sarcoidosis. Clin. Chest Med. 2008, 29, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, Y.; Lim, C.X.; Weichhart, T.; Valeyre, D.; Bentaher, A.; Calender, A. Sarcoidosis and the mTOR, Rac1, and Autophagy Triad. Trends Immunol. 2020, 41, 286–299. [Google Scholar] [CrossRef]
- Mortaz, E.; Adcock, I.M.; Barnes, P.J. Sarcoidosis: Role of non-tuberculosis mycobacteria and Mycobacterium tuberculosis. Int. J. Mycobacteriol. 2014, 3, 225–229. [Google Scholar] [CrossRef]
- Sakhamuru, S.; Kambampati, S.; Wasim, S.; Kukkar, V.; Malik, B.H. The Role of Propionibacterium acnes in the Pathogenesis of Sarcoidosis and Ulcerative Colitis: How This Connection May Inspire Novel Management of These Conditions. Cureus 2020, 12, e10812. [Google Scholar] [CrossRef] [PubMed]
- Gherardi, R.; Coquet, M.; Chérin, P.; Authier, F.-J.; Laforêt, P.; Bélec, L.; Figarella-Branger, D.; Mussini, J.-M.; Pellissier, J.-F.; Fardeau, M. Macrophagic myofasciitis: An emerging entity. Lancet 1998, 352, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Preusse, C.; Goebel, H.-H.; Pehl, D.; Rinnenthal, J.L.; Kley, R.A.; Allenbach, Y.; Heppner, F.L.; Vorgerd, M.; Authier, F.J.; Gherardi, R.; et al. Th2-M2 immunity in lesions of muscular sarcoidosis and macrophagic myofasciitis: Th2-M2 immunity in lesions. Neuropathol. Appl. Neurobiol. 2015, 41, 952–963. [Google Scholar] [CrossRef]
- ten Dam, L.; Raaphorst, J.; van der Kooi, A.J.; Eftimov, F.; Aronica, E.; van de Beek, D.; Brouwer, M.C. Clinical characteristics and outcome in muscular sarcoidosis: A retrospective cohort study and literature review. Neuromuscul. Disord. 2022, 32, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Prieto-González, S.; Grau, J.M. Diagnosis and classification of granulomatous myositis. Autoimmun. Rev. 2014, 13, 372–374. [Google Scholar] [CrossRef]
- Cohen Aubart, F.; Abbara, S.; Maisonobe, T.; Cottin, V.; Papo, T.; Haroche, J.; Mathian, A.; Pha, M.; Gilardin, L.; Hervier, B.; et al. Symptomatic muscular sarcoidosis: Lessons from a nationwide multicenter study. Neurol. Neuroimmunol. Neuroinflamm. 2018, 5, e452. [Google Scholar] [CrossRef] [PubMed]
- Dieudonné, Y.; Allenbach, Y.; Benveniste, O.; Leonard-Louis, S.; Hervier, B.; Mariampillai, K.; Nespola, B.; Lannes, B.; Echaniz-Laguna, A.; Wendling, D.; et al. Granulomatosis-associated myositis: High prevalence of sporadic inclusion body myositis. Neurology 2020, 94, e910–e920. [Google Scholar] [CrossRef] [PubMed]
- Chazaud, B. Inflammation and Skeletal Muscle Regeneration: Leave It to the Macrophages! Trends Immunol. 2020, 41, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Tidball, J.G.; Wehling-Henricks, M. Shifts in macrophage cytokine production drive muscle fibrosis. Nat. Med. 2015, 21, 665–666. [Google Scholar] [CrossRef]
- Coulis, G.; Jaime, D.; Guerrero-Juarez, C.; Kastenschmidt, J.M.; Farahat, P.K.; Nguyen, Q.; Pervolarakis, N.; McLinden, K.; Thurlow, L.; Movahedi, S.; et al. Single-cell and spatial transcriptomics identify a macrophage population associated with skeletal muscle fibrosis. Sci. Adv. 2023, 9, eadd9984. [Google Scholar] [CrossRef]
- Crouser, E.D.; Maier, L.A.; Wilson, K.C.; Bonham, C.A.; Morgenthau, A.S.; Patterson, K.C.; Abston, E.; Bernstein, R.C.; Blankstein, R.; Chen, E.S.; et al. Diagnosis and Detection of Sarcoidosis. An Official American Thoracic Society Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2020, 201, e26–e51. [Google Scholar] [CrossRef]
- Wahlström, J.; Katchar, K.; Wigzell, H.; Olerup, O.; Eklund, A.; Grunewald, J. Analysis of Intracellular Cytokines in CD4+ and CD8+ Lung and Blood T Cells in Sarcoidosis. Am. J. Respir. Crit. Care Med. 2001, 163, 115–121. [Google Scholar] [CrossRef]
- Garret, M.; Pestronk, A. Sarcoidosis, granulomas and myopathy syndromes: A clinical-pathology review. J. Neuroimmunol. 2022, 373, 577975. [Google Scholar] [CrossRef]
- Jaiswal, A.; Rehman, R.; Dutta, J.; Singh, S.; Ray, A.; Shridhar, M.; Jaisankar, J.; Bhatt, M.; Khandelwal, D.; Sahoo, B.; et al. Cellular Distribution of Secreted Phospholipase A2 in Lungs of IPF Patients and Its Inhibition in Bleomycin-Induced Pulmonary Fibrosis in Mice. Cells 2023, 12, 1044. [Google Scholar] [CrossRef]
- Zhang, L.; Du, J.; Hu, Z.; Han, G.; Delafontaine, P.; Garcia, G.; Mitch, W.E. IL-6 and Serum Amyloid A Synergy Mediates Angiotensin II—Induced Muscle Wasting. J. Am. Soc. Nephrol. 2009, 20, 604–612. [Google Scholar] [CrossRef]
- Langhans, C.; Weber-Carstens, S.; Schmidt, F.; Hamati, J.; Kny, M.; Zhu, X.; Wollersheim, T.; Koch, S.; Krebs, M.; Schulz, H.; et al. Inflammation-Induced Acute Phase Response in Skeletal Muscle and Critical Illness Myopathy. PLoS ONE 2014, 9, e92048. [Google Scholar] [CrossRef]
- Panek, R.B.; Moses, H.; Ting, J.P.; Benveniste, E.N. Tumor necrosis factor alpha response elements in the HLA-DRA promoter: Identification of a tumor necrosis factor alpha-induced DNA-protein complex in astrocytes. Proc. Natl. Acad. Sci. USA 1992, 89, 11518–11522. [Google Scholar] [CrossRef] [PubMed]
- Panek, R.B.; Benveniste, E.N. Class II MHC gene expression in microglia. Regulation by the cytokines IFN-gamma, TNF-alpha, and TGF-beta. J. Immunol. Baltim. 1995, 154, 2846–2854. [Google Scholar] [CrossRef]
- Damsky, W.; Thakral, D.; Emeagwali, N.; Galan, A.; King, B. Tofacitinib Treatment and Molecular Analysis of Cutaneous Sarcoidosis. N. Engl. J. Med. 2018, 379, 2540–2546. [Google Scholar] [CrossRef] [PubMed]
- Zuk, J.A.; Fletcher, A. Skeletal muscle expression of class II histocompatibility antigens (HLA-DR) in polymyositis and other muscle disorders with an inflammatory infiltrate. J. Clin. Pathol. 1988, 41, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Englund, P.; Lindroos, E.; Nennesmo, I.; Klareskog, L.; Lundberg, I.E. Skeletal Muscle Fibers Express Major Histocompatibility Complex Class II Antigens Independently of Inflammatory Infiltrates in Inflammatory Myopathies. Am. J. Pathol. 2001, 159, 1263–1273. [Google Scholar] [CrossRef]
- Calvier, L.; Miana, M.; Reboul, P.; Cachofeiro, V.; Martinez-Martinez, E.; De Boer, R.A.; Poirier, F.; Lacolley, P.; Zannad, F.; Rossignol, P.; et al. Galectin-3 Mediates Aldosterone-Induced Vascular Fibrosis. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 67–75. [Google Scholar] [CrossRef]
- Martínez-Martínez, E.; Calvier, L.; Fernández-Celis, A.; Rousseau, E.; Jurado-López, R.; Rossoni, L.V.; Jaisser, F.; Zannad, F.; Rossignol, P.; Cachofeiro, V.; et al. Galectin-3 Blockade Inhibits Cardiac Inflammation and Fibrosis in Experimental Hyperaldosteronism and Hypertension. Hypertension 2015, 66, 767–775. [Google Scholar] [CrossRef]
- Kumagai, K.; Tabu, K.; Sasaki, F.; Takami, Y.; Morinaga, Y.; Mawatari, S.; Hashimoto, S.; Tanoue, S.; Kanmura, S.; Tamai, T.; et al. Glycoprotein Nonmetastatic Melanoma B (Gpnmb)-Positive Macrophages Contribute to the Balance between Fibrosis and Fibrolysis during the Repair of Acute Liver Injury in Mice. PLoS ONE 2015, 10, e0143413. [Google Scholar] [CrossRef]
- Vetrone, S.A.; Montecino-Rodriguez, E.; Kudryashova, E.; Kramerova, I.; Hoffman, E.P.; Liu, S.D.; Miceli, M.C.; Spencer, M.J. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-β. J. Clin. Investig. 2009, 119, 1583–1594. [Google Scholar] [CrossRef]
- Kramerova, I.; Kumagai-Cresse, C.; Ermolova, N.; Mokhonova, E.; Marinov, M.; Capote, J.; Becerra, D.; Quattrocelli, M.; Crosbie, R.H.; Welch, E.; et al. Spp1 (osteopontin) promotes TGFβ processing in fibroblasts of dystrophin-deficient muscles through matrix metalloproteinases. Hum. Mol. Genet. 2019, 28, 3431–3442. [Google Scholar] [CrossRef]
- Franco, C.; Gatto, M.; Iaccarino, L.; Ghirardello, A.; Doria, A. Lymphocyte immunophenotyping in inflammatory myositis: A review. Curr. Opin. Rheumatol. 2021, 33, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Lemos, D.R.; Babaeijandaghi, F.; Low, M.; Chang, C.-K.; Lee, S.T.; Fiore, D.; Zhang, R.-H.; Natarajan, A.; Nedospasov, S.A.; Rossi, F.M.V. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat. Med. 2015, 21, 786–794. [Google Scholar] [CrossRef]
- Piotrowski, W.J.; Kiszałkiewicz, J.; Pastuszak-Lewandoska, D.; Antczak, A.; Górski, P.; Migdalska-Sęk, M.; Górski, W.; Czarnecka, K.; Nawrot, E.; Domańska, D.; et al. TGF-β and SMADs mRNA Expression in Pulmonary Sarcoidosis. In Respiratory Carcinogenesis; Pokorski, M., Ed.; Springer International Publishing: Cham, Switzerland, 2014; Volume 852, pp. 59–69. [Google Scholar]
- Ismaeel, A.; Kim, J.-S.; Kirk, J.S.; Smith, R.S.; Bohannon, W.T.; Koutakis, P. Role of Transforming Growth Factor-β in Skeletal Muscle Fibrosis: A Review. Int. J. Mol. Sci. 2019, 20, 2446. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, I.E.; Eickelberg, O. The Impact of TGF-β on Lung Fibrosis: From Targeting to Biomarkers. Proc. Am. Thorac. Soc. 2012, 9, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Warshamana, G.S.; Corti, M.; Brody, A.R. TNF-α, PDGF, and TGF-β1 Expression by Primary Mouse Bronchiolar-Alveolar Epithelial and Mesenchymal Cells: TNF-α Induces TGF-β1. Exp. Mol. Pathol. 2001, 71, 13–33. [Google Scholar] [CrossRef] [PubMed]
- Theret, M.; Rossi, F.M.V.; Contreras, O. Evolving Roles of Muscle-Resident Fibro-Adipogenic Progenitors in Health, Regeneration, Neuromuscular Disorders, and Aging. Front. Physiol. 2021, 12, 673404. [Google Scholar] [CrossRef]
- Nawaz, A.; Bilal, M.; Fujisaka, S.; Kado, T.; Aslam, M.R.; Ahmed, S.; Okabe, K.; Igarashi, Y.; Watanabe, Y.; Kuwano, T.; et al. Depletion of CD206+ M2-like macrophages induces fibro-adipogenic progenitors activation and muscle regeneration. Nat. Commun. 2022, 13, 7058. [Google Scholar] [CrossRef]
- Prokop, S.; Heppner, F.L.; Goebel, H.H.; Stenzel, W. M2 Polarized Macrophages and Giant Cells Contribute to Myofibrosis in Neuromuscular Sarcoidosis. Am. J. Pathol. 2011, 178, 1279–1286. [Google Scholar] [CrossRef]
- Orandi, A.B.; Eutsler, E.; Ferguson, C.; White, A.J.; Kitcharoensakkul, M. Sarcoidosis presenting as granulomatous myositis in a 16-year-old adolescent. Pediatr. Rheumatol. 2016, 14, 59. [Google Scholar] [CrossRef]
- Mageau, A.; Rigolet, A.; Benali, K.; Chauchard, M.; Ladjeroud, S.; Mahe, I.; Maisonobe, T.; Chauveheid, M.-P.; Papo, T.; Sacre, K. Life-Threatening Hypercalcemia Revealing Diffuse and Isolated Acute Sarcoid-Like Myositis: A New Entity? (A Case-Series). Medicine 2016, 95, e3089. [Google Scholar] [CrossRef]
- Wieërs, G.; Lhommel, R.; Lecouvet, F.; Van Den Bergh, P.; Lambert, M. A tiger man. Lancet 2012, 380, 1859. [Google Scholar] [CrossRef]
- Dalakas, M.C. Inflammatory Muscle Diseases. N. Engl. J. Med. 2015, 372, 1734–1747. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J. Current Classification and Management of Inflammatory Myopathies. J. Neuromuscul. Dis. 2018, 5, 109–129. [Google Scholar] [CrossRef] [PubMed]
- Jamilloux, Y.; Cohen-Aubart, F.; Chapelon-Abric, C.; Maucort-Boulch, D.; Marquet, A.; Pérard, L.; Bouillet, L.; Deroux, A.; Abad, S.; Bielefeld, P.; et al. Efficacy and safety of tumor necrosis factor antagonists in refractory sarcoidosis: A multicenter study of 132 patients. Semin. Arthritis Rheum. 2017, 47, 288–294. [Google Scholar] [CrossRef] [PubMed]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lequain, H.; Dégletagne, C.; Streichenberger, N.; Valantin, J.; Simonet, T.; Schaeffer, L.; Sève, P.; Leblanc, P. Spatial Transcriptomics Reveals Signatures of Histopathological Changes in Muscular Sarcoidosis. Cells 2023, 12, 2747. https://doi.org/10.3390/cells12232747
Lequain H, Dégletagne C, Streichenberger N, Valantin J, Simonet T, Schaeffer L, Sève P, Leblanc P. Spatial Transcriptomics Reveals Signatures of Histopathological Changes in Muscular Sarcoidosis. Cells. 2023; 12(23):2747. https://doi.org/10.3390/cells12232747
Chicago/Turabian StyleLequain, Hippolyte, Cyril Dégletagne, Nathalie Streichenberger, Julie Valantin, Thomas Simonet, Laurent Schaeffer, Pascal Sève, and Pascal Leblanc. 2023. "Spatial Transcriptomics Reveals Signatures of Histopathological Changes in Muscular Sarcoidosis" Cells 12, no. 23: 2747. https://doi.org/10.3390/cells12232747
APA StyleLequain, H., Dégletagne, C., Streichenberger, N., Valantin, J., Simonet, T., Schaeffer, L., Sève, P., & Leblanc, P. (2023). Spatial Transcriptomics Reveals Signatures of Histopathological Changes in Muscular Sarcoidosis. Cells, 12(23), 2747. https://doi.org/10.3390/cells12232747