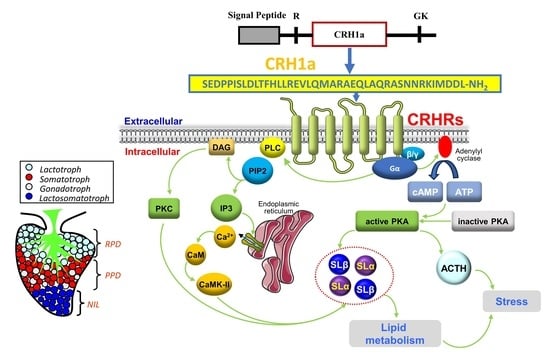
Corticotropin-Releasing Hormone: A Novel Stimulator of Somatolactin in Teleost Pituitary Cells
Abstract
:Highlights
- CRH could significantly induce pituitary somatolactin secretion and synthesis.
- CRH-induced SL expression was mainly mediated by the AC/cAMP/PKA pathway.
- Single-cell transcriptome showed that SLα and SLβ were expressed in different cells of the pituitary NIL region.
- SLα and SLβ could regulate hepatocyte lipid metabolism.
- This study supplemented the function of CRH in the grass carp pituitary.
- This study suggested that there might be another pathway for the CRH response to stress.
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish Acquisition, Acclimation, and Sacrifice
2.2. Chemical Reagents
2.3. Cloning and Tissue Distribution of Grass Carp CRHs and CRHRs
2.4. Transfection and Luciferase Reporter Assay
2.5. SLα and SLβ Secretion and mRNA Expression in Carp Pituitary Cells
2.6. Transcriptome Sequencing and Bioinformatics Analysis
2.7. Immunofluorescence Staining of SLα and SLβ in Grass Carp Pituitary
2.8. Single Cell Transcriptome Sequencing of Grass Carp Pituitary
2.9. Data Transformation and Statistical Analysis
3. Results
3.1. Sequence Analysis of CRHs and CRHRs in Grass Carp
3.2. Transcriptomic Analysis for the Pituitary Actions of CRH1a in Grass Carp
3.3. CRH Could Induce Pituitary Somatolactin Secretion and mRNA Expression
3.4. Signal Transduction for the Regulation of SLα and SLβ by CRH1a
3.5. The Function of SLα and SLβ in Grass Carp Liver
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewis, K.; Li, C.; Perrin, M.H.; Blount, A.; Kunitake, K.; Donaldson, C.; Vaughan, J.; Reyes, T.M.; Gulyas, J.; Fischer, W.; et al. Identification of urocortin III, an additional member of the corticotropin-releasing factor (CRF) family with high affinity for the CRF2 receptor. Proc. Natl. Acad. Sci. USA 2001, 98, 7570–7575. [Google Scholar] [CrossRef] [PubMed]
- Vale, W.; Spiess, J.; Rivier, C.; Rivier, J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science 1981, 213, 1394–1397. [Google Scholar] [CrossRef] [PubMed]
- Spencer, S.J.; Tilbrook, A. The glucocorticoid contribution to obesity. Stress 2011, 14, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Bern, H.A.; Pearson, D.; Larson, B.A.; Nishioka, R.S. Neurohormones from fish tails: The caudal neurosecretory system. I. “Urophysiology” and the caudal neurosecretory system of fishes. Recent Prog. Horm. Res. 1985, 41, 533–552. [Google Scholar] [PubMed]
- Muglia, L.J.; Bethin, K.E.; Jacobson, L.; Vogt, S.K.; Majzoub, J.A. Pituitary-adrenal axis regulation in CRH deficient mice. Endocr. Res. 2000, 26, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Lu, Y.; Zhai, G.; Huang, J.; Shang, G.; Lou, Q.; Li, D.; Jin, X.; He, J.; Du, Z.; et al. Hyperandrogenism in POMCa-deficient zebrafish enhances somatic growth without increasing adiposity. J. Mol. Cell Biol. 2020, 12, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, D.A. Structural evolution of urotensin-I: Reflections of life before corticotropin releasing factor. Gen. Comp. Endocrinol. 2009, 164, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Bern, H.A.; Takasugi, N. The caudal neurosecretory system of fishes. Gen. Comp. Endocrinol. 1962, 2, 96–110. [Google Scholar] [CrossRef]
- Matsuda, K. Recent advances in the regulation of feeding behavior by neuropeptides in fish. Ann. N. Y. Acad. Sci. 2009, 1163, 241–250. [Google Scholar] [CrossRef]
- I Baker, B.; Bird, D.J.; Buckingham, J.C. In the trout, CRH and AVT synergize to stimulate ACTH release. Regul. Pept. 1996, 67, 207–210. [Google Scholar] [CrossRef]
- Ono, M.; Takayama, Y.; Rand-Weaver, M.; Sakata, S.; Yasunaga, T.; Noso, T.; Kawauchi, H. cDNA cloning of somatolactin, a pituitary protein related to growth hormone and prolactin. Proc. Natl. Acad. Sci. USA 1990, 87, 4330–4334. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Stiller, J.; Shaner, M.; Baldini, A.; Scemama, J.; Capehart, A. Cloning of somatolactin alpha and beta cDNAs in zebrafish and phylogenetic analysis of two distinct somatolactin subtypes in fish. J. Endocrinol. 2004, 182, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Benedet, S.; Björnsson, B.T.; Taranger, G.L.; Andersson, E. Cloning of somatolactin alpha, beta forms and the somatolactin receptor in Atlantic salmon: Seasonal expression profile in pituitary and ovary of maturing female broodstock. Reprod. Biol. Endocrinol. 2008, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Ko, W.K.; Lerner, E.A.; Chan, K.M.; Wong, A.O. Grass carp somatolactin: I. Evidence for PACAP induction of somatolactin-alpha and -beta gene expression via activation of pituitary PAC-I receptors. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E463–E476. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Song, D.; Tran, N.T.; Nguyen, N. The effects of the members of growth hormone family knockdown in zebrafish development. Gen. Comp. Endocrinol. 2007, 150, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Fukamachi, S.; Asakawa, S.; Wakamatsu, Y.; Shimizu, N.; Mitani, H.; Shima, A. Conserved function of medaka pink-eyed dilution in melanin synthesis and its divergent transcriptional regulation in gonads among vertebrates. Genetics 2004, 168, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wong, A.O.L. Signal transduction mechanisms for autocrine/paracrine regulation of somatolactin α secretion and synthesis in carp pituitary cells by somatolactin α and β. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E176–E186. [Google Scholar] [CrossRef] [PubMed]
- Sasano, Y.; Yoshimura, A.; Fukamachi, S. Reassessment of the function of somatolactin alpha in lipid metabolism using medaka mutant and transgenic strains. BMC Genet. 2012, 13, 64. [Google Scholar] [CrossRef]
- Sudo, R.; Suetake, H.; Suzuki, Y.; Aoyama, J.; Tsukamoto, K. Profiles of mRNA expression for prolactin, growth hormone, and somatolactin in Japanese eels, Anguilla japonica: The effect of salinity, silvering and seasonal change. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2013, 164, 10–16. [Google Scholar] [CrossRef]
- Rand-Weaver, M.; Pottinger, T.G.; Sumpter, J.P. Plasma somatolactin concentrations in salmonid fish are elevated by stress. J. Endocrinol. 1993, 138, 509–515. [Google Scholar] [CrossRef]
- Khalil, N.A.; Hashem, A.M.; Ibrahim, A.A.E.; Mousa, M.A. Effect of stress during handling, seawater acclimation, confinement, and induced spawning on plasma ion levels and somatolactin-expressing cells in mature female Liza ramada. J. Exp. Zool. A Ecol. Genet. Physiol. 2012, 317, 410–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.H.; Xie, B.W.; Wang, Z.J.; Zhang, Y.G. Characterizaiton and expression analysis of somatolactin-α and –β genes in rare minnows (Gobiocypris rarus) following waterborne cadmium exposure. Fish Physiol. Biochem. 2018, 44, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Marco, M.; Gao, Y.L.; Wong, M.H. Sustainable management of non-native grass carp as a protein source, weed-control agent and sport fish. Aquac. Res. 2022, 53, 5809–5824. [Google Scholar] [CrossRef]
- Hu, G.; He, M.; Ko, W.K.; Lin, C.; Wong, A.O. Novel pituitary actions of TAC3 gene products in fish model: Receptor specificity and signal transduction for prolactin and somatolactin alpha regulation by neurokinin B (NKB) and NKB-related peptide in carp pituitary cells. Endocrinology 2014, 155, 3582–3596. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.O.; Ng, S.; Lee, E.K.; Leung, R.C.; Ho, W.K. Somatostatin inhibits (D-Arg6, Pro9-NEt) salmon gonadotropin-releasing hormone- and dopamine D1- stimulated growth hormone release from perifused pituitary cells of Chinese grass carp, Ctenopharyngodon idellus. Gen. Comp. Endocrinol. 1998, 110, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Qin, Q.; Xu, S.; Zhou, L.; Xia, C.; Shi, X.; Zhang, H.; Jia, J.; Ye, C.; Yin, Z.; et al. Pituitary Actions of EGF on Gonadotropins, Growth Hormone, Prolactin and Somatolactins in Grass Carp. Biology 2020, 9, 279. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Qin, X.; Zhou, L.; Shi, X.; Cai, T.; Xie, Y.; Li, W.; Du, R.; OuYang, Y.; Yin, Z.; et al. Reproductive Regulation of PrRPs in Teleost: The Link Between Feeding and Reproduction. Front. Endocrinol. 2021, 12, 762826. [Google Scholar] [CrossRef]
- Im, K.; Mareninov, S.; Diaz, M.F.P.; Yong, W.H. An Introduction to Performing Immunofluorescence Staining. Methods Mol. Biol. 2019, 1897, 299–311. [Google Scholar]
- Siddique, K.; Ager-Wick, E.; Fontaine, R.; Weltzien, F.-A.; Henkel, C.V. Characterization of hormone-producing cell types in the teleost pituitary gland using single-cell RNA-seq. Sci. Data 2021, 8, 279. [Google Scholar] [CrossRef]
- Hillhouse, E.W.; Grammatopoulos, D.K. The molecular mechanisms underlying the regulation of the biological activity of corticotropin-releasing hormone receptors: Implications for physiology and pathophysiology. Endocr. Rev. 2006, 27, 260–286. [Google Scholar] [CrossRef]
- Gysling, K.; Forray, M.I.; Haeger, P.; Daza, C.; Rojas, R. Corticotropin-releasing hormone and urocortin: Redundant or distinctive functions? Brain Res. Brain Res. Rev. 2004, 47, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.H.; Lowry, C.A. Corticotropin-releasing factor-related peptides, serotonergic systems, and emotional behavior. Front. Neurosci. 2013, 7, 169. [Google Scholar] [CrossRef] [PubMed]
- Vetter, D.E.; Li, C.; Zhao, L.; Contarino, A.; Liberman, M.C.; Smith, G.W.; Marchuk, Y.; Koob, G.F.; Heinemann, S.F.; Vale, W.; et al. Urocortin-deficient mice show hearing impairment and increased anxiety-like behavior. Nat. Genet. 2002, 31, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Lee, J.H.; Yoon, Y.; Pleasure, S.J.; Yoon, K.J. The CRHR1/CREB/REST signaling cascade regulates mammalian embryonic neural stem cell properties. EMBO Rep. 2022, 24, e55313. [Google Scholar] [CrossRef] [PubMed]
- Onuma, T.; Ando, H.; Koide, N.; Okada, H.; Urano, A. Effects of salmon GnRH and sex steroid hormones on expression of genes encoding growth hormone/rolactin/somatolactin family hormones and a pituitary-specific transcription factor in masu salmon pituitary cells in vitro. Gen. Comp. Endocinol. 2005, 143, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Rhee, J.S.; Kim, B.M.; Seo, J.S.; Kim, I.C.; Lee, Y.M.; Lee, J.S. Cloning of growth hormone, somatolacin and their receptor mRNAs, their expression in organs, during development, and on salinity stress in the hermaphroditic fish, Kryptolebias marmoratus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2012, 161, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Fukada, H.; Ozaki, Y.; Pierce, A.L.; Adachi, S.; Yamauchi, K.; Hara, A.; Swanso, P.; Dickhoff, W.W. Identification of the salmon somatolactin receptor, a new member of the cytokine receptor family. Endocrinology 2005, 146, 2354–2361. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Ward, W.F. PGC-1alpha: A key regulator of energy metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef]
- Cheng, C.-F.; Ku, H.-C.; Lin, H. PGC-1α as a pivotal factor in lipid and metabolic regulation. Int. J. Mol. Sci. 2018, 19, 3447. [Google Scholar] [CrossRef]
- Peng, X.; Shang, G.; Wang, W.; Chen, X.; Lou, Q.; Zhai, G.; Li, D.; Du, Z.; Ye, Y.; Jin, X. Fatty acid oxidation in zebrafish adipose tissue is promoted by 1α,25(OH)2D3. Cell Rep. 2017, 19, 1444–1455. [Google Scholar] [CrossRef]
- Deng, W.; Sun, J.; Chang, Z.-G.; Gou, N.-N.; Wu, W.-Y.; Luo, X.-L.; Zhou, J.-S.; Yu, H.-B.; Ji, H. Energy response and fatty acid metabolism in Onychostoma macrolepis exposed to low-temperature stress. J. Therm. Biol. 2020, 94, 102725. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, R.; Shi, X.; Chen, F.; Wang, L.; Liang, H.; Hu, G. Corticotropin-Releasing Hormone: A Novel Stimulator of Somatolactin in Teleost Pituitary Cells. Cells 2023, 12, 2770. https://doi.org/10.3390/cells12242770
Du R, Shi X, Chen F, Wang L, Liang H, Hu G. Corticotropin-Releasing Hormone: A Novel Stimulator of Somatolactin in Teleost Pituitary Cells. Cells. 2023; 12(24):2770. https://doi.org/10.3390/cells12242770
Chicago/Turabian StyleDu, Ruixin, Xuetao Shi, Feng Chen, Li Wang, Hongwei Liang, and Guangfu Hu. 2023. "Corticotropin-Releasing Hormone: A Novel Stimulator of Somatolactin in Teleost Pituitary Cells" Cells 12, no. 24: 2770. https://doi.org/10.3390/cells12242770
APA StyleDu, R., Shi, X., Chen, F., Wang, L., Liang, H., & Hu, G. (2023). Corticotropin-Releasing Hormone: A Novel Stimulator of Somatolactin in Teleost Pituitary Cells. Cells, 12(24), 2770. https://doi.org/10.3390/cells12242770