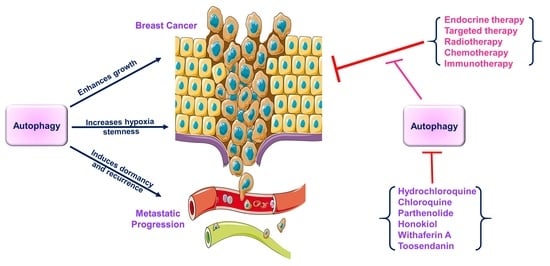
Autophagy and Breast Cancer: Connected in Growth, Progression, and Therapy
Abstract
:1. Autophagy, a Complex Process Designed to Support Cell Death as Well as Survival via Recycling
2. Complex Relationship between Key Autophagic Proteins and Various Aspects of Breast Cancer Growth
2.1. BECN1 Negatively Associates with Breast Cancer
2.2. Autophagy-Related Genes (ATGs) Have a Dual Impact on Breast Cancer
2.3. FOXO Can Modulate Breast Cancer via Autophagy
2.4. Autophagy Contributes to Reduced Drug Sensitivity in Breast Cancer
2.5. Autophagy Influences Tumor Dormancy in Breast Cancer
2.6. Autophagy Influences Hypoxia, Chemoresistance, and Stem-like Phenotype in Breast Cancer
2.7. Intermediate Steps in the Autophagic Process Play an Important Role in Breast Cancer
3. A multifaceted Involvement of Autophagic Processes Modulate the Efficacy of Breast Cancer Therapeutics
3.1. Autophagy and Endocrine Therapy—Response and Resistance
3.2. Autophagy and Targeted Therapy—Response and Resistance
3.3. Autophagy and Radiotherapy—Response and Resistance
3.4. Autophagy and Chemotherapy—Response and Resistance
3.5. Autophagy and Immunotherapy—Response and Resistance
4. Inhibition of Autophagy Using Various Inhibitors May Result in Improved Therapeutic Outcomes
4.1. Hydroxychloroquine (HCQ)
4.2. Chloroquine (CQ)
4.3. Parthenolide
4.4. Honokiol
4.5. Withaferin A
4.6. Toosendanin
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bednarczyk, M.; Zmarzły, N.; Grabarek, B.; Mazurek, U.; Muc-Wierzgoń, M. Genes involved in the regulation of different types of autophagy and their participation in cancer pathogenesis. Oncotarget 2018, 9, 34413–34428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deretic, V. Autophagy in inflammation, infection, and immunometabolism. Immunity 2021, 54, 437–453. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef]
- Guan, J.-L.; Simon, A.K.; Prescott, M.; Menendez, J.A.; Liu, F.; Wang, F.; Wang, C.; Wolvetang, E.; Vazquez-Martin, A.; Zhang, J. Autophagy in stem cells. Autophagy 2013, 9, 830–849. [Google Scholar] [CrossRef]
- Chang, N.C. Autophagy and Stem Cells: Self-Eating for Self-Renewal. Front. Cell Dev. Biol. 2020, 8, 138. [Google Scholar] [CrossRef] [Green Version]
- Galluzzi, L.; Green, D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef]
- Jia, S.-N.; Lin, C.; Chen, D.-F.; Li, A.-Q.; Dai, L.; Zhang, L.; Zhao, L.-L.; Yang, J.-S.; Yang, F.; Yang, W.-J. The Transcription Factor p8 Regulates Autophagy in Response to Palmitic Acid Stress via a Mammalian Target of Rapamycin (mTOR)-independent Signaling Pathway. J. Biol. Chem. 2016, 291, 4462–4472. [Google Scholar] [CrossRef] [Green Version]
- Parzych, K.R.; Klionsky, D.J. An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid. Redox Signal. 2014, 20, 460–473. [Google Scholar] [CrossRef] [Green Version]
- Mancias, J.D.; Kimmelman, A.C. Mechanisms of Selective Autophagy in Normal Physiology and Cancer. J. Mol. Biol. 2016, 428, 1659–1680. [Google Scholar] [CrossRef] [Green Version]
- Conway, O.; Akpinar, H.A.; Rogov, V.V.; Kirkin, V. Selective Autophagy Receptors in Neuronal Health and Disease. J. Mol. Biol. 2020, 432, 2483–2509. [Google Scholar] [CrossRef] [PubMed]
- Wirawan, E.; Vanden Berghe, T.; Lippens, S.; Agostinis, P.; Vandenabeele, P. Autophagy: For better or for worse. Cell Res. 2012, 22, 43–61. [Google Scholar] [CrossRef] [Green Version]
- Mijaljica, D.; Prescott, M.; Devenish, R.J. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy 2011, 7, 673–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massey, A.; Kiffin, R.; Cuervo, A.M. Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2420–2434. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Klionsky, D.J. Autophagy: Molecular machinery for self-eating. Cell Death Differ. 2005, 12 (Suppl. S2), 1542–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.J.; Ye, L.; Huang, W.F.; Guo, L.J.; Xu, Z.G.; Wu, H.L.; Yang, C.; Liu, H.F. p62 links the autophagy pathway and the ubiqutin–proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 2016, 21, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faruk, M.O.; Ichimura, Y.; Komatsu, M. Selective autophagy. Cancer Sci. 2021, 112, 3972–3978. [Google Scholar] [CrossRef]
- Münch, C.; Dikic, I. Hitchhiking on selective autophagy. Nat. Cell Biol. 2018, 20, 122–124. [Google Scholar] [CrossRef]
- Xiong, H.; Shen, J.; Chen, Z.; Yang, J.; Xie, B.; Jia, Y.; Jayasinghe, U.; Wang, J.; Zhao, W.; Xie, S.; et al. H19/let-7/Lin28 ceRNA network mediates autophagy inhibiting epithelial-mesenchymal transition in breast cancer. Int. J. Oncol. 2020, 56, 794–806. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, B.; Zhu, H.; Qu, X.; Zhao, L.; Tan, Y.; Jiang, Y.; Liao, M.; Wu, X. Inhibition of long non-coding RNA ROR reverses resistance to Tamoxifen by inducing autophagy in breast cancer. Tumor Biol. 2017, 39, 1010428317705790. [Google Scholar] [CrossRef] [Green Version]
- Qian, X.; Qu, H.; Zhang, F.; Peng, S.; Dou, D.; Yang, Y.; Ding, Y.; Xie, M.; Dong, H.; Liao, Y.; et al. Exosomal long noncoding RNA AGAP2-AS1 regulates trastuzumab resistance via inducing autophagy in breast cancer. Am. J. Cancer Res. 2021, 11, 1962–1981. [Google Scholar] [PubMed]
- Han, X.; Mo, J.; Yang, Y.; Wang, Y.; Lu, H. Crucial Roles of LncRNAs-Mediated Autophagy in Breast Cancer. Int. J. Med. Sci. 2022, 19, 1082–1092. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jin, F.; Li, Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J. Cell. Mol. Med. 2021, 25, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Hewitt, G.; Korolchuk, V.I. Repair, Reuse, Recycle: The Expanding Role of Autophagy in Genome Maintenance. Trends Cell Biol. 2017, 27, 340–351. [Google Scholar] [CrossRef] [Green Version]
- Galluzzi, L.; Pietrocola, F.; Bravo-San Pedro, J.M.; Amaravadi, R.K.; Baehrecke, E.H.; Cecconi, F.; Codogno, P.; Debnath, J.; Gewirtz, D.A.; Karantza, V.; et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015, 34, 856–880. [Google Scholar] [CrossRef] [PubMed]
- Rybstein, M.D.; Bravo-San Pedro, J.M.; Kroemer, G.; Galluzzi, L. The autophagic network and cancer. Nat. Cell Biol. 2018, 20, 243–251. [Google Scholar] [CrossRef]
- Young, A.R.; Narita, M.; Ferreira, M.; Kirschner, K.; Sadaie, M.; Darot, J.F.; Tavaré, S.; Arakawa, S.; Shimizu, S.; Watt, F.M.; et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009, 23, 798–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santana-Codina, N.; Mancias, J.D.; Kimmelman, A.C. The Role of Autophagy in Cancer. Annu. Rev. Cancer Biol. 2017, 1, 19–39. [Google Scholar] [CrossRef]
- Maslov, A.Y.; Vijg, J. Genome instability, cancer and aging. Biochim. Biophys. Acta 2009, 1790, 963–969. [Google Scholar] [CrossRef] [Green Version]
- Garufi, A.; Pucci, D.; D’Orazi, V.; Cirone, M.; Bossi, G.; Avantaggiati, M.L.; D’Orazi, G. Degradation of mutant p53H175 protein by Zn(II) through autophagy. Cell Death Dis. 2014, 5, e1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choundhury, S.; Kolukula, V.K.; Preet, A.; Albanese, C.; Avantaggiati, M.L. Dissecting the pathways that destabilize mutant p53: The proteasome or autophagy? Cell Cycle 2013, 12, 1022–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, O.C.; Choudhury, S.; Kolukula, V.; Vietsch, E.E.; Catania, J.; Preet, A.; Reynoso, K.; Bargonetti, J.; Wellstein, A.; Albanese, C.; et al. Dietary downregulation of mutant p53 levels via glucose restriction. Cell Cycle 2012, 11, 4436–4446. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Chitiprolu, M.; Gagnon, D.; Meng, L.; Perez-Iratxeta, C.; Lagace, D.; Gibbings, D. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat. Commun. 2014, 5, 5276. [Google Scholar] [CrossRef] [Green Version]
- Rello-Varona, S.; Lissa, D.; Shen, S.; Niso-Santano, M.; Senovilla, L.; Mariño, G.; Vitale, I.; Jemaa, M.; Harper, F.; Pierron, G.; et al. Autophagic removal of micronuclei. Cell Cycle 2012, 11, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Karp, C.M.; Beaudoin, B.; Vuong, N.; Chen, G.; Chen, H.-Y.; Bray, K.; Reddy, A.; Bhanot, G.; Gelinas, C.; et al. Autophagy Suppresses Tumorigenesis through Elimination of p62. Cell 2009, 137, 1062–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.-S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.-I.; Ezaki, J.; Murata, S.; et al. Homeostatic Levels of p62 Control Cytoplasmic Inclusion Body Formation in Autophagy-Deficient Mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Hori, T.; Cooper, T.K.; Liao, J.; Desai, N.; Serfass, J.M.; Young, M.M.; Park, S.; Izu, Y.; Wang, H.-G. Bif-1 haploinsufficiency promotes chromosomal instability and accelerates Myc-driven lymphomagenesis via suppression of mitophagy. Blood 2013, 121, 1622–1632. [Google Scholar] [CrossRef] [Green Version]
- Green, D.R.; Galluzzi, L.; Kroemer, G. Mitochondria and the Autophagy-Inflammation-Cell Death Axis in Organismal Aging. Science 2011, 333, 1109–1112. [Google Scholar] [CrossRef] [Green Version]
- Greim, H.; Kaden, D.A.; Larson, R.A.; Palermo, C.M.; Rice, J.M.; Ross, D.; Snyder, R. The bone marrow niche, stem cells, and leukemia: Impact of drugs, chemicals, and the environment. Ann. N. Y. Acad. Sci. 2014, 1310, 7–31. [Google Scholar] [CrossRef] [Green Version]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Autophagy and Cellular Immune Responses. Immunity 2013, 39, 211–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niklaus, N.J.; Tokarchuk, I.; Zbinden, M.; Schläfli, A.M.; Maycotte, P.; Tschan, M.P. The Multifaceted Functions of Autophagy in Breast Cancer Development and Treatment. Cells 2021, 10, 1447. [Google Scholar] [CrossRef] [PubMed]
- Tavera-Mendoza, L.E.; Westerling, T.; Libby, E.; Marusyk, A.; Cato, L.; Cassani, R.; Cameron, L.A.; Ficarro, S.B.; Marto, J.A.; Klawitter, J.; et al. Vitamin D receptor regulates autophagy in the normal mammary gland and in luminal breast cancer cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2186–E2194. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef]
- Miracco, C.; Cosci, E.; Oliveri, G.; Luzi, P.; Pacenti, L.; Monciatti, I.; Mannucci, S.; De Nisi, M.C.; Toscano, M.; Malagnino, V.; et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int. J. Oncol. 2007, 30, 429–436. [Google Scholar]
- Vega-Rubin-de-Celis, S.; Zou, Z.; Fernandez, A.F.; Ci, B.; Kim, M.; Xiao, G.; Xie, Y.; Levine, B. Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc. Natl. Acad. Sci. USA 2018, 115, 4176–4181. [Google Scholar] [CrossRef] [Green Version]
- Cicchini, M.; Chakrabarti, R.; Kongara, S.; Price, S.; Nahar, R.; Lozy, F.; Zhong, H.; Vazquez, A.; Kang, Y.; Karantza, V. Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy 2014, 10, 2036–2052. [Google Scholar] [CrossRef] [Green Version]
- Hatzis, C.; Pusztai, L.; Valero, V.; Booser, D.J.; Esserman, L.; Lluch, A.; Vidaurre, T.; Holmes, F.; Souchon, E.; Wang, H.; et al. A Genomic Predictor of Response and Survival Following Taxane-Anthracycline Chemotherapy for Invasive Breast Cancer. JAMA 2011, 305, 1873–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuli, M.V.; Giuliani, E.; Screpanti, I.; Bellavia, D.; Checquolo, S. Notch Signaling Activation as a Hallmark for Triple-Negative Breast Cancer Subtype. J. Oncol. 2019, 2019, 8707053. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.-S.; Ann, E.-J.; Kim, M.-Y.; Yoon, J.-H.; Lee, H.-J.; Jo, E.-H.; Lee, K.; Lee, J.S.; Park, H.-S. Autophagy negatively regulates tumor cell proliferation through phosphorylation dependent degradation of the Notch1 intracellular domain. Oncotarget 2016, 7, 79047–79063. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, J.; Li, S.; Feng, Y.; Yi, F.; Wang, L.; Wei, S.; Cao, L. Autophagy-related 7 modulates tumor progression in triple-negative breast cancer. Lab. Investig. 2019, 99, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Lock, R.; Roy, S.; Kenific, C.M.; Su, J.S.; Salas, E.; Ronen, S.M.; Debnath, J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell 2011, 22, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Maycotte, P.; Gearheart, C.M.; Barnard, R.; Aryal, S.; Mulcahy Levy, J.M.; Fosmire, S.P.; Hansen, R.J.; Morgan, M.J.; Porter, C.C.; Gustafson, D.L.; et al. STAT3-Mediated Autophagy Dependence Identifies Subtypes of Breast Cancer Where Autophagy Inhibition Can Be Efficacious. Cancer Res. 2014, 74, 2579–2590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornsveld, M.; Smits, L.M.M.; Meerlo, M.; van Amersfoort, M.; Groot Koerkamp, M.J.A.; van Leenen, D.; Kloet, D.E.A.; Holstege, F.C.P.; Derksen, P.W.B.; Burgering, B.M.T.; et al. FOXO Transcription Factors Both Suppress and Support Breast Cancer Progression. Cancer Res. 2018, 78, 2356–2369. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Elshimali, Y.; Sarkissyan, M.; Mohamed, H.; Clayton, S.; Vadgama, J.V. Expression of FOXO1 is associated with GATA3 and Annexin-1 and predicts disease-free survival in breast cancer. Am. J. Cancer Res. 2012, 2, 104–115. [Google Scholar] [CrossRef]
- Habashy, H.O.; Rakha, E.A.; Aleskandarany, M.; Ahmed, M.A.; Green, A.; Ellis, I.O.; Powe, D.G. FOXO3a nuclear localisation is associated with good prognosis in luminal-like breast cancer. Breast Cancer Res. Treat. 2011, 129, 11–21. [Google Scholar] [CrossRef]
- Smit, L.; Berns, K.; Spence, K.; Ryder, W.D.; Zeps, N.; Madiredjo, M.; Beijersbergen, R.; Bernards, R.; Clarke, R.B. An integrated genomic approach identifies that the PI3K/AKT/FOXO pathway is involved in breast cancer tumor initiation. Oncotarget 2016, 7, 2596–2610. [Google Scholar] [CrossRef] [Green Version]
- Schäffner, I.; Minakaki, G.; Khan, M.A.; Balta, E.-A.; Schlötzer-Schrehardt, U.; Schwarz, T.J.; Beckervordersandforth, R.; Winner, B.; Webb, A.E.; DePinho, R.A.; et al. FoxO Function Is Essential for Maintenance of Autophagic Flux and Neuronal Morphogenesis in Adult Neurogenesis. Neuron 2018, 99, 1188–1203.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audesse, A.J.; Dhakal, S.; Hassell, L.-A.; Gardell, Z.; Nemtsova, Y.; Webb, A.E. FOXO3 directly regulates an autophagy network to functionally regulate proteostasis in adult neural stem cells. PLoS Genet. 2019, 15, e1008097. [Google Scholar] [CrossRef] [PubMed]
- Sharif, T.; Martell, E.; Dai, C.; Kennedy, B.E.; Murphy, P.; Clements, D.R.; Kim, Y.; Lee, P.W.K.; Gujar, S.A. Autophagic homeostasis is required for the pluripotency of cancer stem cells. Autophagy 2017, 13, 264–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boya, P.; Codogno, P.; Rodriguez-Muela, N. Autophagy in stem cells: Repair, remodelling and metabolic reprogramming. Development 2018, 145, dev146506. [Google Scholar] [CrossRef] [Green Version]
- Azamjah, N.; Soltan-Zadeh, Y.; Zayeri, F. Global Trend of Breast Cancer Mortality Rate: A 25-Year Study. Asian Pac. J. Cancer Prev. 2019, 20, 2015–2020. [Google Scholar] [CrossRef]
- Vera-Ramirez, L.; Vodnala, S.K.; Nini, R.; Hunter, K.W.; Green, J.E. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat. Commun. 2018, 9, 1944. [Google Scholar] [CrossRef] [Green Version]
- Maishman, T.; Cutress, R.I.; Hernandez, A.; Gerty, S.; Copson, E.R.; Durcan, L.; Eccles, D.M. Local Recurrence and Breast Oncological Surgery in Young Women With Breast Cancer: The POSH Observational Cohort Study. Ann. Surg. 2017, 266, 165–172. [Google Scholar] [CrossRef] [Green Version]
- Clarke, S.J.; Rivory, L. Clinical Pharmacokinetics of Docetaxel. Clin. Pharmacokinet. 1999, 36, 99–114. [Google Scholar] [CrossRef]
- Trudeau, M.; Charbonneau, F.; Gelmon, K.; Laing, K.; Latreille, J.; Mackey, J.; McLeod, D.; Pritchard, K.; Provencher, L.; Verma, S. Selection of adjuvant chemotherapy for treatment of node-positive breast cancer. Lancet Oncol. 2005, 6, 886–898. [Google Scholar] [CrossRef]
- Jamieson, E.R.; Lippard, S.J. Structure, recognition, and processing of cisplatin—DNA adducts. Chem. Rev. 1999, 99, 2467–2498. [Google Scholar] [CrossRef]
- Thomas, S.S.; Clements, P.J.; Furst, D.E. Immunosuppressives: Cyclosporine, cyclophosphamide, azathioprine, mycophenolate mofetil. In Rheumatology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 459–467. [Google Scholar]
- Wen, N.; Lv, Q.; Du, Z.-G. MicroRNAs involved in drug resistance of breast cancer by regulating autophagy. J. Zhejiang Univ. Sci. B 2020, 21, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Rossari, F.; Zucchinetti, C.; Buda, G.; Orciuolo, E. Tumor dormancy as an alternative step in the development of chemoresistance and metastasis-clinical implications. Cell. Oncol. 2020, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, G.G.; Deshmukh, A.P.; den Hollander, P.; Luo, M.; Soundararajan, R.; Jia, D.; Levine, H.; Mani, S.A.; Wicha, M.S. Breast cancer dormancy: Need for clinically relevant models to address current gaps in knowledge. NPJ Breast Cancer 2021, 7, 66. [Google Scholar] [CrossRef]
- Sosa, M.S.; Bragado, P.; Aguirre-Ghiso, J.A. Mechanisms of disseminated cancer cell dormancy: An awakening field. Nat. Rev. Cancer 2014, 14, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Katheder, N.S.; Khezri, R.; O’Farrell, F.; Schultz, S.W.; Jain, A.; Rahman, M.M.; Schink, K.O.; Theodossiou, T.A.; Johansen, T.; Juhász, G.; et al. Microenvironmental autophagy promotes tumour growth. Nature 2017, 541, 417–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaravadi, R.; Kimmelman, A.C.; White, E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016, 30, 1913–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.G.; MacLeod, K.F. Autophagy, cancer stem cells and drug resistance. J. Pathol. 2019, 247, 708–718. [Google Scholar] [CrossRef] [Green Version]
- La Belle Flynn, A.; Calhoun, B.C.; Sharma, A.; Chang, J.C.; Almasan, A.; Schiemann, W.P. Autophagy inhibition elicits emergence from metastatic dormancy by inducing and stabilizing Pfkfb3 expression. Nat. Commun. 2019, 10, 3668. [Google Scholar] [CrossRef] [Green Version]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [Green Version]
- Settembre, C.; Fraldi, A.; Medina, D.L.; Ballabio, A. Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013, 14, 283–296. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.C.; Guan, K.-L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Investig. 2015, 125, 25–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldman, M.E.; Apsel, B.; Uotila, A.; Loewith, R.; Knight, Z.A.; Ruggero, D.; Shokat, K.M. Active-Site Inhibitors of mTOR Target Rapamycin-Resistant Outputs of mTORC1 and mTORC2. PLoS Biol. 2009, 7, e38. [Google Scholar] [CrossRef]
- Marinković, M.; Šprung, M.; Buljubasic, M.; Novak, I. Autophagy Modulation in Cancer: Current Knowledge on Action and Therapy. Oxidative Med. Cell. Longev. 2018, 2018, 8023821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Luo, R.Z.; Lu, Y.; Zhang, X.; Yu, Q.; Khare, S.; Kondo, S.; Kondo, Y.; Yu, Y.; Mills, G.B.; et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J. Clin. Investig. 2008, 118, 3917–3929. [Google Scholar] [CrossRef] [Green Version]
- Zou, C.-F.; Jia, L.; Jin, H.; Yao, M.; Zhao, N.; Huan, J.; Lu, Z.; Bast, R.C., Jr.; Feng, Y.; Yu, Y. Re-expression of ARHI (DIRAS3) induces autophagy in breast cancer cells and enhances the inhibitory effect of paclitaxel. BMC Cancer 2011, 11, 22. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Triple-negative breast cancer therapeutic resistance: Where is the Achilles’ heel? Cancer Lett. 2021, 497, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Cancer stem cell in breast cancer therapeutic resistance. Cancer Treat. Rev. 2018, 69, 152–163. [Google Scholar] [CrossRef]
- Lee, K.-M.; Giltnane, J.M.; Balko, J.M.; Schwarz, L.J.; Guerrero-Zotano, A.L.; Hutchinson, K.E.; Nixon, M.J.; Estrada, M.V.; Sánchez, V.; Sanders, M.E.; et al. MYC and MCL1 Cooperatively Promote Chemotherapy-Resistant Breast Cancer Stem Cells via Regulation of Mitochondrial Oxidative Phosphorylation. Cell Metab. 2017, 26, 633–647.e7. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Settleman, J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 2010, 29, 4741–4751. [Google Scholar] [CrossRef] [Green Version]
- Nazio, F.; Bordi, M.; Cianfanelli, V.; Locatelli, F.; Cecconi, F. Autophagy and cancer stem cells: Molecular mechanisms and therapeutic applications. Cell Death Differ. 2019, 26, 690–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bousquet, G.; El Bouchtaoui, M.; Sophie, T.; Leboeuf, C.; de Bazelaire, C.; Ratajczak, P.; Giacchetti, S.; de Roquancourt, A.; Bertheau, P.; Verneuil, L.; et al. Targeting autophagic cancer stem-cells to reverse chemoresistance in human triple negative breast cancer. Oncotarget 2017, 8, 35205–35221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, R.; Karantza-Wadsworth, V.; White, E. Role of autophagy in cancer. Nat. Rev. Cancer 2007, 7, 961–967. [Google Scholar] [CrossRef]
- Thomas, M.; Davis, T.; Loos, B.; Sishi, B.; Huisamen, B.; Strijdom, H.; Engelbrecht, A.-M. Autophagy is essential for the maintenance of amino acids and ATP levels during acute amino acid starvation in MDAMB231 cells. Cell Biochem. Funct. 2018, 36, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Dudek-Peric, A.M.; Garg, A.D.; Roose, H.; Demirsoy, S.; Van Eygen, S.; Mertens, F.; Vangheluwe, P.; Vankelecom, H.; Agostinis, P. An autophagy-driven pathway of ATP secretion supports the aggressive phenotype of BRAF(V600E) inhibitor-resistant metastatic melanoma cells. Autophagy 2017, 13, 1512–1527. [Google Scholar] [CrossRef] [Green Version]
- Schito, L.; Semenza, G.L. Hypoxia-Inducible Factors: Master Regulators of Cancer Progression. Trends Cancer 2016, 2, 758–770. [Google Scholar] [CrossRef] [Green Version]
- Michiels, C. Physiological and Pathological Responses to Hypoxia. Am. J. Pathol. 2004, 164, 1875–1882. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Salnikov, A.V.; Bauer, N.; Aleksandrowicz, E.; Labsch, S.; Nwaeburu, C.; Mattern, J.; Gladkich, J.; Schemmer, P.; Werner, J.; et al. Triptolide reverses hypoxia-induced epithelial–mesenchymal transition and stem-like features in pancreatic cancer by NF-κB downregulation. Int. J. Cancer 2014, 134, 2489–2503. [Google Scholar] [CrossRef] [Green Version]
- Samanta, D.; Gilkes, D.M.; Chaturvedi, P.; Xiang, L.; Semenza, G.L. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, E5429–E5438. [Google Scholar] [CrossRef] [Green Version]
- Daskalaki, I.; Gkikas, I.; Tavernarakis, N. Hypoxia and Selective Autophagy in Cancer Development and Therapy. Front. Cell Dev. Biol. 2018, 6, 104. [Google Scholar] [CrossRef] [Green Version]
- Yeo, S.K.; Wen, J.; Chen, S.; Guan, J.-L. Autophagy Differentially Regulates Distinct Breast Cancer Stem-like Cells in Murine Models via EGFR/Stat3 and Tgfβ/Smad Signaling. Cancer Res. 2016, 76, 3397–3410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golden, E.B.; Cho, H.-Y.; Jahanian, A.; Hofman, F.M.; Louie, S.G.; Schönthal, A.H.; Chen, T.C. Chloroquine enhances temozolomide cytotoxicity in malignant gliomas by blocking autophagy. Neurosurg. Focus 2014, 37, E12. [Google Scholar] [CrossRef] [Green Version]
- Chaterjee, M.; Van Golen, K.L. Breast Cancer Stem Cells Survive Periods of Farnesyl-Transferase Inhibitor-Induced Dormancy by Undergoing Autophagy. Bone Marrow Res. 2011, 2011, 362938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Xu, L.; Zhang, F.; Vlashi, E. Doxycycline inhibits the cancer stem cell phenotype and epithelial-to-mesenchymal transition in breast cancer. Cell Cycle 2017, 16, 737–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibragimova, M.; Tsyganov, M.; Litviakov, N. Tumour Stem Cells in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 5058. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Martin, A.; Oliveras-Ferraros, C.; Menendez, J.A. Autophagy Facilitates the Development of Breast Cancer Resistance to the Anti-HER2 Monoclonal Antibody Trastuzumab. PLoS ONE 2009, 4, e6251. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.-L.; Chen, J.; Wang, Y.-P.; Zheng, H. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy 2011, 7, 1035–1044. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Tian, L.; Jung, M.; Choi, S.K.; Sun, Y.; Kim, H.; Moon, W.K. Downregulation of Choline Kinase-Alpha Enhances Autophagy in Tamoxifen-Resistant Breast Cancer Cells. PLoS ONE 2015, 10, e0141110. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Chen, T.; Li, G.; Xu, C.; Xu, Z.; Zhang, J.; He, K.; Zheng, L.; Guan, Z.; Su, X.; et al. Lower Beclin 1 downregulates HER2 expression to enhance tamoxifen sensitivity and predicts a favorable outcome for ER positive breast cancer. Oncotarget 2017, 8, 52156–52177. [Google Scholar] [CrossRef] [Green Version]
- Berardi, D.E.; Flumian, C.; Rodriguez, C.E.; Bessone, M.I.; Cirigliano, S.M.; Joffé, E.D.; Fiszman, G.L.; Urtreger, A.J.; Todaro, L.B. PKCδ Inhibition Impairs Mammary Cancer Proliferative Capacity But Selects Cancer Stem Cells, Involving Autophagy. J. Cell. Biochem. 2016, 117, 730–740. [Google Scholar] [CrossRef]
- Xiang, S.; Dauchy, R.T.; Hoffman, A.E.; Pointer, D.; Frasch, T.; Blask, D.E.; Hill, S.M. Epigenetic inhibition of the tumor suppressor ARHI by light at night-induced circadian melatonin disruption mediates STAT3-driven paclitaxel resistance in breast cancer. J. Pineal Res. 2019, 67, e12586. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.M.; Blask, D.E.; Xiang, S.; Yuan, L.; Mao, L.; Dauchy, R.T.; Dauchy, E.M.; Frasch, T.; Duplesis, T. Melatonin and Associated Signaling Pathways that Control Normal Breast Epithelium and Breast Cancer. J. Mammary Gland. Biol. Neoplasia 2011, 16, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Shen, S.; Zhang, Y.J.; Xu, C.-F.; Cao, Z.-T.; Wen, L.-P.; Wang, J. Nanoparticle-facilitated autophagy inhibition promotes the efficacy of chemotherapeutics against breast cancer stem cells. Biomaterials 2016, 103, 44–55. [Google Scholar] [CrossRef]
- Liang, D.H.; Choi, D.S.; Ensor, J.E.; Kaipparettu, B.A.; Bass, B.L.; Chang, J.C. The autophagy inhibitor chloroquine targets cancer stem cells in triple negative breast cancer by inducing mitochondrial damage and impairing DNA break repair. Cancer Lett. 2016, 376, 249–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, W.; Hamaï, A.; Tonelli, G.; Bauvy, C.; Nicolas, V.; Tharinger, H.; Codogno, P.; Mehrpour, M. Inhibition of the autophagic flux by salinomycin in breast cancer stem-like/progenitor cells interferes with their maintenance. Autophagy 2013, 9, 714–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Periyasamy-Thandavan, S.; Jackson, W.H.; Samaddar, J.S.; Erickson, B.; Barrett, J.R.; Raney, L.; Gopal, E.; Ganapathy, V.; Hill, W.D.; Bhalla, K.N.; et al. Bortezomib blocks the catabolic process of autophagy via a cathepsin-dependent mechanism, affects endoplasmic reticulum stress, and induces caspase-dependent cell death in antiestrogen–sensitive and resistant ER+ breast cancer cells. Autophagy 2010, 6, 19–35. [Google Scholar] [CrossRef] [Green Version]
- Schwartz-Roberts, J.L.; Shajahan, A.N.; Cook, K.L.; Wärri, A.; Abu-Asab, M.; Clarke, R. GX15-070 (Obatoclax) Induces Apoptosis and Inhibits Cathepsin D- and L–Mediated Autophagosomal Lysis in Antiestrogen-Resistant Breast Cancer Cells. Mol. Cancer Ther. 2013, 12, 448–459. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zou, L.; Li, Q.; Haibe-Kains, B.; Tian, R.; Li, Y.; Desmedt, C.; Sotiriou, C.; Szallasi, Z.; Iglehart, J.D.; et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat. Med. 2010, 16, 214–218. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, Q.; Tian, R.; Wang, Q.; Zhao, J.J.; Iglehart, J.D.; Wang, Z.C.; Richardson, A.L. Lysosomal Transmembrane Protein LAPTM4B Promotes Autophagy and Tolerance to Metabolic Stress in Cancer Cells. Cancer Res. 2011, 71, 7481–7489. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Iglehart, J.D.; Richardson, A.L.; Wang, Z.C. The amplified cancer gene LAPTM4B promotes tumor growth and tolerance to stress through the induction of autophagy. Autophagy 2012, 8, 273–274. [Google Scholar] [CrossRef] [Green Version]
- Heng, M.Y.; Detloff, P.J.; Paulson, H.L.; Albin, R.L. Early alterations of autophagy in Huntington disease-like mice. Autophagy 2010, 6, 1206–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decressac, M.; Mattsson, B.; Weikop, P.; Lundblad, M.; Jakobsson, J.; Bjorklund, A. TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl. Acad. Sci. USA 2013, 110, E1817–E1826. [Google Scholar] [CrossRef] [Green Version]
- Kimura, T.; Takabatake, Y.; Takahashi, A.; Isaka, Y. Chloroquine in Cancer Therapy: A Double-Edged Sword of Autophagy. Cancer Res. 2013, 73, 3–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.-J.; Lei, Y.-H.; Yao, N.; Wang, C.-R.; Hu, N.; Ye, W.-C.; Zhang, D.-M.; Chen, Z.-S. Autophagy and multidrug resistance in cancer. Chin. J. Cancer 2017, 36, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, B.; Turner, N.C.; Jones, A. An update on the medical management of breast cancer. BMJ 2014, 348, g3608. [Google Scholar] [CrossRef]
- Davies, C.; Godwin, J.; Gray, R.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Pan, H.C.; Taylor, C.; Wang, Y.C.; et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar] [CrossRef] [Green Version]
- Nass, N.; Kalinski, T. Tamoxifen resistance: From cell culture experiments towards novel biomarkers. Pathol. Res. Pract. 2015, 211, 189–197. [Google Scholar] [CrossRef]
- Dixon, J.M. Endocrine Resistance in Breast Cancer. New J. Sci. 2014, 2014, 390618. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Blum, J.; Yang, X.; Beaulieu, N.; MacLeod, A.R.; Davidson, N.E. Release of Methyl CpG Binding Proteins and Histone Deacetylase 1 from the Estrogen Receptor α (ER) Promoter upon Reactivation in ER-Negative Human Breast Cancer Cells. Mol. Endocrinol. 2005, 19, 1740–1751. [Google Scholar] [CrossRef] [Green Version]
- Jeselsohn, R.; Buchwalter, G.; DE Angelis, C.; Brown, M.; Schiff, R. ESR1 mutations—A mechanism for acquired endocrine resistance in breast cancer. Nat. Rev. Clin. Oncol. 2015, 12, 573–583. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Liu, S.; Luo, H.; Chen, C.; Zhang, X.; He, L.; Tu, G. GPR30-mediated HMGB1 upregulation in CAFs induces autophagy and tamoxifen resistance in ERα-positive breast cancer cells. Aging 2021, 13, 16178–16197. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 2019, 12, 81. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.; Liu, M.; Yang, F.; Luo, H.; Li, Z.; Tu, G.; Yang, G. GPR30 as an initiator of tamoxifen resistance in hormone-dependent breast cancer. Breast Cancer Res. 2013, 15, R114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lappano, R.; De Marco, P.; De Francesco, E.M.; Chimento, A.; Pezzi, V.; Maggiolini, M. Cross-talk between GPER and growth factor signaling. J. Steroid Biochem. Mol. Biol. 2013, 137, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Yang, G.; Hou, Y.; Tang, X.; Wu, C.; Wu, X.-A.; Guo, L.; Zhu, Q.; Luo, H.; Du, Y.-E.; et al. Cytoplasmic GPER translocation in cancer-associated fibroblasts mediates cAMP/PKA/CREB/glycolytic axis to confer tumor cells with multidrug resistance. Oncogene 2017, 36, 2131–2145. [Google Scholar] [CrossRef] [PubMed]
- Henson, E.S.; Gibson, S.B. Surviving cell death through epidermal growth factor (EGF) signal transduction pathways: Implications for cancer therapy. Cell. Signal. 2006, 18, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Yang, H.; Wang, G.; Sun, L.; Zhou, Y.; Guo, Y.; Xi, Z.; Jiang, X. Autophagy augmented by troglitazone is independent of EGFR transactivation and correlated with AMP-activated protein kinase signaling. Autophagy 2010, 6, 67–73. [Google Scholar] [CrossRef] [Green Version]
- Madeo, A.; Maggiolini, M. Nuclear Alternate Estrogen Receptor GPR30 Mediates 17β-Estradiol–Induced Gene Expression and Migration in Breast Cancer–Associated Fibroblasts. Cancer Res. 2010, 70, 6036–6046. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Yang, G.; Yu, T.; Luo, S.; Wu, C.; Sun, Y.; Liu, M.; Tu, G. GPER-mediated proliferation and estradiol production in breast cancer-associated fibroblasts. Endocr. Relat. Cancer 2014, 21, 355–369. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.-H.; Koh, D.; Na, H.; Ka, N.-L.; Kim, S.; Kim, H.-J.; Hong, S.; Shin, Y.K.; Seong, J.K.; Lee, M.-O. MTA1 is a novel regulator of autophagy that induces tamoxifen resistance in breast cancer cells. Autophagy 2018, 14, 812–824. [Google Scholar] [CrossRef]
- Desta, Z.; Ward, B.A.; Soukhova, N.V.; Flockhart, D.A. Comprehensive Evaluation of Tamoxifen Sequential Biotransformation by the Human Cytochrome P450 System in Vitro: Prominent Roles for CYP3A and CYP2D6. J. Pharmacol. Exp. Ther. 2004, 310, 1062–1075. [Google Scholar] [CrossRef]
- Shajahan-Haq, A.N.; Cook, K.L.; Schwartz-Roberts, J.L.; Eltayeb, A.E.; Demas, D.M.; Warri, A.M.; Facey, C.O.; Hilakivi-Clarke, L.A.; Clarke, R. MYC regulates the unfolded protein response and glucose and glutamine uptake in endocrine resistant breast cancer. Mol. Cancer 2014, 13, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, K.L.; Clarke, R. Role of GRP78 in promoting therapeutic-resistant breast cancer. Future Med. Chem. 2015, 7, 1529–1534. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.L.; Shajahan, A.N.; Wärri, A.; Jin, L.; Hilakivi-Clarke, L.A.; Clarke, R. Glucose-Regulated Protein 78 Controls Cross-talk between Apoptosis and Autophagy to Determine Antiestrogen Responsiveness. Cancer Res. 2012, 72, 3337–3349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, K.L.; Clarke, P.A.G.; Parmar, J.; Hu, R.; Schwartz-Roberts, J.L.; Abu-Asab, M.; Wärri, A.; Baumann, W.T.; Clarke, R. Knockdown of estrogen receptor-α induces autophagy and inhibits antiestrogen-mediated unfolded protein response activation, promoting ROS-induced breast cancer cell death. FASEB J. 2014, 28, 3891–3905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, R.; Tyson, J.J.; Dixon, J.M. Endocrine resistance in breast cancer—An overview and update. Mol. Cell. Endocrinol. 2015, 418, 220–234. [Google Scholar] [CrossRef] [Green Version]
- Clarke, R.; Cook, K.L.; Hu, R.; Facey, C.O.; Tavassoly, I.; Schwartz, J.L.; Baumann, W.T.; Tyson, J.J.; Xuan, J.; Wang, Y.; et al. Endoplasmic Reticulum Stress, the Unfolded Protein Response, Autophagy, and the Integrated Regulation of Breast Cancer Cell Fate. Cancer Res. 2012, 72, 1321–1331. [Google Scholar] [CrossRef] [Green Version]
- Grkovic, S.; O’Reilly, V.C.; Han, S.; Hong, M.; Baxter, R.C.; Firth, S.M. IGFBP-3 binds GRP78, stimulates autophagy and promotes the survival of breast cancer cells exposed to adverse microenvironments. Oncogene 2013, 32, 2412–2420. [Google Scholar] [CrossRef] [Green Version]
- Gomez, B.P.; Riggins, R.B.; Shajahan, A.N.; Klimach, U.; Wang, A.; Crawford, A.C.; Zhu, Y.; Zwart, A.; Wang, M.; Clarke, R. Human X-Box binding protein-1 confers both estrogen independence and antiestrogen resistance in breast cancer cell lines. FASEB J. 2007, 21, 4013–4027. [Google Scholar] [CrossRef]
- Crawford, A.C.; Riggins, R.B.; Shajahan, A.N.; Zwart, A.; Clarke, R. Co-Inhibition of BCL-W and BCL2 Restores Antiestrogen Sensitivity through BECN1 and Promotes an Autophagy-Associated Necrosis. PLoS ONE 2010, 5, e8604. [Google Scholar] [CrossRef] [Green Version]
- Prensner, J.R.; Chinnaiyan, A.M. The Emergence of lncRNAs in Cancer Biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Luo, A.; Liu, Y.; Wang, S.; Li, Y.; Shi, W.; Liu, Z.; Qu, X. MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 2015, 14, 208. [Google Scholar] [CrossRef] [Green Version]
- Jung, C.H.; Ro, S.-H.; Cao, J.; Otto, N.M.; Kim, D.-H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.-X.; Pan, H.; Ouyang, D.-Y.; He, X.-H. The critical molecular interconnections in regulating apoptosis and autophagy. Ann. Med. 2015, 47, 305–315. [Google Scholar] [CrossRef]
- Schumacker, P.T. Reactive oxygen species in cancer cells: Live by the sword, die by the sword. Cancer Cell 2006, 10, 175–176. [Google Scholar] [CrossRef] [Green Version]
- Bacci, M.; Lorito, N.; Ippolito, L.; Ramazzotti, M.; Luti, S.; Romagnoli, S.; Parri, M.; Bianchini, F.; Cappellesso, F.; Virga, F.; et al. Reprogramming of Amino Acid Transporters to Support Aspartate and Glutamate Dependency Sustains Endocrine Resistance in Breast Cancer. Cell Rep. 2019, 28, 104–118.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meijer, A.J.; Lorin, S.; Blommaart, E.F.; Codogno, P. Regulation of autophagy by amino acids and MTOR-dependent signal transduction. Amino Acids 2015, 47, 2037–2063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sikder, M.O.F.; Sivaprakasam, S.; Brown, T.P.; Thangaraju, M.; Bhutia, Y.D.; Ganapathy, V. SLC6A14, a Na+/Cl−-coupled amino acid transporter, functions as a tumor promoter in colon and is a target for Wnt signaling. Biochem. J. 2020, 477, 1409–1425. [Google Scholar] [CrossRef] [Green Version]
- Ziaeemehr, A.; Shahidsales, S.; Ghosi, Z.; Avan, A.; Aldavood, A.S.; Anvari, K.; Makhdoomi, Y.; Asadi, M. Association of the involvement of axillary lymph nodes in HER-2/neu overexpression in patients with breast cancer. Breast J. 2019, 25, 537–538. [Google Scholar] [CrossRef]
- Giannone, G.; Milani, A.; Geuna, E.; Galizia, D.; Biello, F.; Montemurro, F. What is the best pharmacotherapeutic strategy for HER-2 positive breast cancer? Expert Opin. Pharmacother. 2019, 20, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, A.; Kruczak, A.; Harazin-Lechowska, A.; Ambicka, A.; Grela-Wojewoda, A.; Domagała-Haduch, M.; Janecka-Widła, A.; Majchrzyk, K.; Cichocka, A.; Ryś, J.; et al. Relationship between HER2 gene status and selected potential biological features related to trastuzumab resistance and its influence on survival of breast cancer patients undergoing trastuzumab adjuvant treatment. OncoTargets Ther. 2018, 11, 4525–4535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datta, A.; Loo, S.Y.; Huang, B.; Wong, L.; Tan, S.S.; Tan, T.Z.; Lee, S.-C.; Thiery, J.P.; Lim, Y.C.; Yong, W.P.; et al. SPHK1 regulates proliferation and survival responses in triple-negative breast cancer. Oncotarget 2014, 5, 5920–5933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maczis, M.A.; Maceyka, M.; Waters, M.R.; Newton, J.; Singh, M.; Rigsby, M.F.; Turner, T.H.; Alzubi, M.A.; Harrell, J.C.; Milstien, S.; et al. Sphingosine kinase 1 activation by estrogen receptor α36 contributes to tamoxifen resistance in breast cancer. J. Lipid Res. 2018, 59, 2297–2307. [Google Scholar] [CrossRef] [Green Version]
- Carreras, I.; Aytan, N.; Choi, J.-K.; Tognoni, C.M.; Kowall, N.W.; Jenkins, B.G.; Dedeoglu, A. Dual dose-dependent effects of fingolimod in a mouse model of Alzheimer’s disease. Sci. Rep. 2019, 9, 10972. [Google Scholar] [CrossRef] [Green Version]
- White, C.; Alshaker, H.; Cooper, C.; Winkler, M.; Pchejetski, D. The emerging role of FTY720 (Fingolimod) in cancer treatment. Oncotarget 2016, 7, 23106–23127. [Google Scholar] [CrossRef] [Green Version]
- Chung, W.P.; Huang, W.L.; Liao, W.A.; Hung, C.H.; Chiang, C.W.; Cheung, C.H.A.; Su, W.C. FTY720 in resistant human epidermal growth factor receptor 2-positive breast cancer. Sci. Rep. 2022, 12, 241. [Google Scholar] [CrossRef]
- Kataoka, Y.; Mukohara, T.; Shimada, H.; Saijo, N.; Hirai, M.; Minami, H. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann. Oncol. 2010, 21, 255–262. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Øvervatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.-H.; Ampuero, J.; Gil-Gómez, A.; Montero-Vallejo, R.; Rojas, Á.; Muñoz-Hernández, R.; Gallego-Durán, R.; Romero-Gómez, M. miRNAs in patients with non-alcoholic fatty liver disease: A systematic review and meta-analysis. J. Hepatol. 2018, 69, 1335–1348. [Google Scholar] [CrossRef]
- Yu, Y.; Chai, J. The function of miRNAs and their potential as therapeutic targets in burn-induced insulin resistance (Review). Int. J. Mol. Med. 2015, 35, 305–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.; Hu, J.; Lu, P.; Cao, H.; Yu, C.; Li, X.; Qian, X.; Yang, X.; Yang, Y.; Han, N.; et al. Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell Death Dis. 2020, 11, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.; Qian, X.; Cao, H.; Wang, F.; Li, X.; Han, N.; Yang, X.; Yang, Y.; Dou, D.; Hu, J.; et al. lncRNA ZNF649-AS1 Induces Trastuzumab Resistance by Promoting ATG5 Expression and Autophagy. Mol. Ther. 2020, 28, 2488–2502. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Appert-Collin, A.; Hubert, P.; Crémel, G.; Bennasroune, A. Role of ErbB Receptors in Cancer Cell Migration and Invasion. Front. Pharmacol. 2015, 6, 283. [Google Scholar] [CrossRef] [Green Version]
- Tsang, R.Y.; Sadeghi, S.; Finn, R.S. Lapatinib, a Dual-Targeted Small Molecule Inhibitor of EGFR and HER2, in HER2-Amplified Breast Cancer: From Bench to Bedside. Clin. Med. Insights Ther. 2011, 3, CMT.S3783. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Li, X.; Feng, J.; Chang, Y.; Wang, Z.; Wen, A. Autophagy facilitates the Lapatinib resistance of HER2 positive breast cancer cells. Med. Hypotheses 2011, 77, 206–208. [Google Scholar] [CrossRef]
- Huang, M.; Zhou, Y.; Duan, D.; Yang, C.; Zhou, Z.; Li, F.; Kong, Y.; Hsieh, Y.-C.; Zhang, R.; Ding, W.; et al. Targeting ubiquitin conjugating enzyme UbcH5b by a triterpenoid PC3-15 from Schisandra plants sensitizes triple-negative breast cancer cells to lapatinib. Cancer Lett. 2021, 504, 125–136. [Google Scholar] [CrossRef]
- Moulder, S.L.; Yakes, F.M.; Muthuswamy, S.K.; Bianco, R.; Simpson, J.F.; Arteaga, C.L. Epidermal growth factor receptor (HER1) tyrosine kinase inhibitor ZD1839 (Iressa) inhibits HER2/neu (erbB2)-overexpressing breast cancer cells in vitro and in vivo. Cancer Res. 2001, 61, 8887–8895. [Google Scholar]
- Anido, J.; Matar, P.; Albanell, J.; Guzmán, M.; Rojo, F.; Arribas, J.; Averbuch, S.; Baselga, J. ZD1839, a specific epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor, induces the formation of inactive EGFR/HER2 and EGFR/HER3 heterodimers and prevents heregulin signaling in HER2-overexpressing breast cancer cells. Clin. Cancer Res. 2003, 9, 1274–1283. [Google Scholar]
- Campiglio, M.; Locatelli, A.; Olgiati, C.; Normanno, N.; Somenzi, G.; Viganò, L.; Fumagalli, M.; Ménard, S.; Gianni, L. Inhibition of proliferation and induction of apoptosis in breast cancer cells by the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor ZD1839 (‘Iressa’) is independent of EGFR expression level. J. Cell. Physiol. 2004, 198, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Albanell, J.; Ruiz, A.; Lluch, A.; Gascón, P.; Guillém, V.; González, S.; Sauleda, S.; Marimón, I.; Tabernero, J.M.; et al. Phase II and Tumor Pharmacodynamic Study of Gefitinib in Patients with Advanced Breast Cancer. J. Clin. Oncol. 2005, 23, 5323–5333. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, T.O.; Hsu, F.D.; Jensen, K.; Cheang, M.; Karaca, G.; Hu, Z.; Hernandez-Boussard, T.; Livasy, C.; Cowan, D.; Dressler, L.; et al. Immunohistochemical and Clinical Characterization of the Basal-Like Subtype of Invasive Breast Carcinoma. Clin. Cancer Res. 2004, 10, 5367–5374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, W.; Sun, H.; Zhao, Y.; Chen, M.; Yang, X.; Liu, Y.; Jin, W. BCAP31 drives TNBC development by modulating ligand-independent EGFR trafficking and spontaneous EGFR phosphorylation. Theranostics 2019, 9, 6468–6484. [Google Scholar] [CrossRef]
- Liao, M.; Qin, R.; Huang, W.; Zhu, H.-P.; Peng, F.; Han, B.; Liu, B. Targeting regulated cell death (RCD) with small-molecule compounds in triple-negative breast cancer: A revisited perspective from molecular mechanisms to targeted therapies. J. Hematol. Oncol. 2022, 15, 44. [Google Scholar] [CrossRef]
- Liu, Z.; He, K.; Ma, Q.; Yu, Q.; Liu, C.; Ndege, I.; Wang, X.; Yu, Z. Autophagy inhibitor facilitates gefitinib sensitivity in vitro and in vivo by activating mitochondrial apoptosis in triple negative breast cancer. PLoS ONE 2017, 12, e0177694. [Google Scholar] [CrossRef] [Green Version]
- Dragowska, W.H.; Weppler, S.A.; Wang, J.C.; Wong, L.Y.; Kapanen, A.I.; Rawji, J.S.; Warburton, C.; Qadir, M.A.; Donohue, E.; Roberge, M.; et al. Induction of Autophagy Is an Early Response to Gefitinib and a Potential Therapeutic Target in Breast Cancer. PLoS ONE 2013, 8, e76503. [Google Scholar] [CrossRef] [Green Version]
- Weihua, Z.; Tsan, R.; Huang, W.-C.; Wu, Q.; Chiu, C.-H.; Fidler, I.J.; Hung, M.-C. Survival of Cancer Cells Is Maintained by EGFR Independent of Its Kinase Activity. Cancer Cell 2008, 13, 385–393. [Google Scholar] [CrossRef] [Green Version]
- Pernas, S.; Tolaney, S.M.; Winer, E.P.; Goel, S. CDK4/6 inhibition in breast cancer: Current practice and future directions. Ther. Adv. Med. Oncol. 2018, 10, 1758835918786451. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Geng, Y.; Sicinski, P. Specific protection against breast cancers by cyclin D1 ablation. Nature 2001, 411, 1017–1021. [Google Scholar] [CrossRef]
- Arnold, A.; Papanikolaou, A. Cyclin D1 in Breast Cancer Pathogenesis. J. Clin. Oncol. 2005, 23, 4215–4224. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Sicinska, E.; Geng, Y.; Ahnström, M.; Zagozdzon, A.; Kong, Y.; Gardner, H.; Kiyokawa, H.; Harris, L.N.; Stål, O.; et al. Requirement for CDK4 kinase function in breast cancer. Cancer Cell 2006, 9, 23–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casimiro, M.C.; Di Sante, G.; Di Rocco, A.; Loro, E.; Pupo, C.; Pestell, T.G.; Bisetto, S.; Velasco-Velázquez, M.A.; Jiao, X.; Li, Z.; et al. Cyclin D1 Restrains Oncogene-Induced Autophagy by Regulating the AMPK–LKB1 Signaling Axis. Cancer Res. 2017, 77, 3391–3405. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Chen, Y. AMPK and Autophagy. Adv. Exp. Med. Biol. 2019, 1206, 85–108. [Google Scholar] [CrossRef] [PubMed]
- Piezzo, M.; Chiodini, P.; Riemma, M.; Cocco, S.; Caputo, R.; Cianniello, D.; Di Gioia, G.; Di Lauro, V.; Rella, F.D.; Fusco, G.; et al. Progression-Free Survival and Overall Survival of CDK 4/6 Inhibitors Plus Endocrine Therapy in Metastatic Breast Cancer: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2020, 21, 6400. [Google Scholar] [CrossRef]
- Piezzo, M.; Cocco, S.; Caputo, R.; Cianniello, D.; Gioia, G.D.; Lauro, V.D.; Fusco, G.; Martinelli, C.; Nuzzo, F.; Pensabene, M.; et al. Targeting Cell Cycle in Breast Cancer: CDK4/6 Inhibitors. Int. J. Mol. Sci. 2020, 21, 6479. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Q.; Peng, X.; Zhou, C.; Zhong, Y.; Chen, X.; Qiu, Y.; Jin, M.; Gong, M.; Kong, D. Stellettin B Induces G1 Arrest, Apoptosis and Autophagy in Human Non-small Cell Lung Cancer A549 Cells via Blocking PI3K/Akt/mTOR Pathway. Sci. Rep. 2016, 6, 27071. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.J.; Chee, C.E.; Huang, S.; Sinicrope, F.A. The Role of Autophagy in Cancer: Therapeutic Implications. Mol. Cancer Ther. 2011, 10, 1533–1541. [Google Scholar] [CrossRef] [Green Version]
- Vijayaraghavan, S.; Karakas, C.; Doostan, I.; Chen, X.; Bui, T.; Yi, M.; Raghavendra, A.S.; Zhao, Y.; Bashour, S.I.; Ibrahim, N.K.; et al. CDK4/6 and autophagy inhibitors synergistically induce senescence in Rb positive cytoplasmic cyclin E negative cancers. Nat. Commun. 2017, 8, 15916. [Google Scholar] [CrossRef] [Green Version]
- Nardone, V.; Barbarino, M.; Angrisani, A.; Correale, P.; Pastina, P.; Cappabianca, S.; Reginelli, A.; Mutti, L.; Miracco, C.; Giannicola, R.; et al. CDK4, CDK6/cyclin-D1 Complex Inhibition and Radiotherapy for Cancer Control: A Role for Autophagy. Int. J. Mol. Sci. 2021, 22, 8391. [Google Scholar] [CrossRef]
- Cocco, S.; Leone, A.; Piezzo, M.; Caputo, R.; Di Lauro, V.; Di Rella, F.; Fusco, G.; Capozzi, M.; Gioia, G.D.; Budillon, A.; et al. Targeting Autophagy in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 7836. [Google Scholar] [CrossRef] [PubMed]
- Lanceta, L.; O’Neill, C.; Lypova, N.; Li, X.; Rouchka, E.; Waigel, S.; Gomez-Gutierrez, J.G.; Chesney, J.; Imbert-Fernandez, Y. Transcriptomic Profiling Identifies Differentially Expressed Genes in Palbociclib-Resistant ER+ MCF7 Breast Cancer Cells. Genes 2020, 11, 467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fassl, A.; Brain, C.; Abu-Remaileh, M.; Stukan, I.; Butter, D.; Stepien, P.; Feit, A.S.; Bergholz, J.; Michowski, W.; Otto, T.; et al. Increased lysosomal biomass is responsible for the resistance of triple-negative breast cancers to CDK4/6 inhibition. Sci. Adv. 2020, 6, eabb2210. [Google Scholar] [CrossRef] [PubMed]
- Scheidemann, E.R.; Shajahan-Haq, A.N. Resistance to CDK4/6 Inhibitors in Estrogen Receptor-Positive Breast Cancer. Int. J. Mol. Sci. 2021, 22, 12292. [Google Scholar] [CrossRef]
- Connell, P.P.; Hellman, S. Advances in Radiotherapy and Implications for the Next Century: A Historical Perspective. Cancer Res. 2009, 69, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Tam, S.Y.; Wu, V.W.C.; Law, H.K.W. Influence of autophagy on the efficacy of radiotherapy. Radiat. Oncol. 2017, 12, 57. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.; Zhu, Z.; Wu, J.; She, H.; Han, R.; Xu, H.; Qin, Z.-H. DRAM1 regulates autophagy and cell proliferation via inhibition of the phosphoinositide 3-kinase-Akt-mTOR-ribosomal protein S6 pathway. Cell Commun. Signal. 2019, 17, 28. [Google Scholar] [CrossRef] [Green Version]
- Meng, C.; Liu, Y.; Shen, Y.; Liu, S.; Wang, Z.; Ye, Q.; Liu, H.; Liu, X.; Jia, L. MicroRNA-26b suppresses autophagy in breast cancer cells by targeting DRAM1 mRNA, and is downregulated by irradiation. Oncol. Lett. 2018, 15, 1435–1440. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Zhang, W.; Cui, Z.; Chen, Q.; Xie, P.; Zhou, C.; Liu, B.; Peng, X.; Zhang, Y. High mobility group box-1 and its clinical value in breast cancer. OncoTargets Ther. 2015, 8, 413–419. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.Y.; Arnott, D.; Brown, E.J. Ubiquilin4 is an adaptor protein that recruits Ubiquilin1 to the autophagy machinery. EMBO Rep. 2013, 14, 373–381. [Google Scholar] [CrossRef]
- Ahmad, A. Introduction to Cancer Metastasis; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Tang, D.; Kang, R.; Zeh, H.J., III; Lotze, M.T. High-mobility group box 1 and cancer. Biochim. Biophys. Acta 2010, 1799, 131–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.Q.; Huang, B.F.; Wang, Y.; Hu, G.R.; Wang, Q.; Shao, J.K. Expression of HMGB1 protein in breast cancer and its clinicopathological significance. Zhonghua Bing Li Xue Za Zhi 2020, 49, 57–61. [Google Scholar] [PubMed]
- Kang, R.; Livesey, K.M.; Zeh, I.H.J.; Loze, M.T.; Tang, D. HMGB1: A novel Beclin 1-binding protein active in autophagy. Autophagy 2010, 6, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Apetoh, L.; Ghiringhelli, F.; Tesniere, A.; Criollo, A.; Ortiz, C.; Lidereau, R.; Mariette, C.; Chaput, N.; Mira, J.-P.; Delaloge, S.; et al. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunol. Rev. 2007, 220, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Ni, P.; Zhang, Y.; Liu, Y.; Lin, X.; Su, X.; Lu, H.; Shen, H.; Xu, W.; Xu, H.; Su, Z. HMGB1 silence could promote MCF-7 cell apoptosis and inhibit invasion and metastasis. Int. J. Clin. Exp. Pathol. 2015, 8, 15940–15946. [Google Scholar] [PubMed]
- Wang, L.-L.; Meng, Q.-H.; Jiao, Y.; Xu, J.-Y.; Ge, C.-M.; Zhou, J.-Y.; Rosen, E.M.; Wang, H.-C.; Fan, S.-J. High-Mobility Group Boxes Mediate Cell Proliferation and Radiosensitivity via Retinoblastoma-Interaction-Dependent and -Independent Mechanisms. Cancer Biother. Radiopharm. 2012, 27, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Qi, X.; Zhang, X.; Gao, D.; Fang, K.; Guo, Z.; Li, L. Med19 is involved in chemoresistance by mediating autophagy through HMGB1 in breast cancer. J. Cell. Biochem. 2019, 120, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Wang, H.-C.; Fan, S.-J. Growth suppression and radiosensitivity increase by HMGB1 in breast cancer. Acta Pharmacol. Sin. 2007, 28, 1957–1967. [Google Scholar] [CrossRef] [Green Version]
- Ke, S.; Zhou, F.; Yang, H.; Wei, Y.; Gong, J.; Mei, Z.; Wu, L.; Yu, H.; Zhou, Y. Downregulation of high mobility group box 1 modulates telomere homeostasis and increases the radiosensitivity of human breast cancer cells. Int. J. Oncol. 2015, 46, 1051–1058. [Google Scholar] [CrossRef] [Green Version]
- Ladoire, S.; Enot, D.; Senovilla, L.; Ghiringhelli, F.; Poirier-Colame, V.; Chaba, K.; Semeraro, M.; Chaix, M.; Penault-Llorca, F.; Arnould, L.; et al. The presence of LC3B puncta and HMGB1 expression in malignant cells correlate with the immune infiltrate in breast cancer. Autophagy 2016, 12, 864–875. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Chen, J.; He, L. mir-129-5p Attenuates Irradiation-Induced Autophagy and Decreases Radioresistance of Breast Cancer Cells by Targeting HMGB1. Med. Sci. Monit. 2015, 21, 4122–4129. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.; Chen, Z.; Huang, J.; Chen, J.; Yuan, W.; Deng, Z.; Chen, Z. Upregulation of Autophagy-Related Gene-5 (ATG-5) Is Associated with Chemoresistance in Human Gastric Cancer. PLoS ONE 2014, 9, e110293. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Li, Y.; Wang, S.; Wang, X.; Liao, C.; Hu, X.; Fan, L.; Kang, Q.; Zeng, Y.; Wu, X.; et al. Inhibition of autophagosome-lysosome fusion by ginsenoside Ro via the ESR2-NCF1-ROS pathway sensitizes esophageal cancer cells to 5-fluorouracil-induced cell death via the CHEK1-mediated DNA damage checkpoint. Autophagy 2016, 12, 1593–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chittaranjan, S.; Bortnik, S.; Dragowska, W.H.; Xu, J.; Abeysundara, N.; Leung, A.; Go, N.E.; DeVorkin, L.; Weppler, S.A.; Gelmon, K.; et al. Autophagy Inhibition Augments the Anticancer Effects of Epirubicin Treatment in Anthracycline-Sensitive and -Resistant Triple-Negative Breast Cancer. Clin. Cancer Res. 2014, 20, 3159–3173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.; Duan, J.-J.; Bian, X.-W.; Yu, S.-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Ghersi, D.; Wilcken, N.; Simes, R.J. A systematic review of taxane-containing regimens for metastatic breast cancer. Br. J. Cancer 2005, 93, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Siddharth, S.; Sharma, D. Triple Negative Breast Cancer: A Mountain Yet to Be Scaled Despite the Triumphs. Cancers 2021, 13, 3697. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Kuei, C.-H.; Lee, H.-H.; Lin, C.-H.; Chen, Y.-L.; Chen, C.-L.; Lin, Y.-F. TNFSF13 upregulation confers chemotherapeutic resistance via triggering autophagy initiation in triple-negative breast cancer. J. Mol. Med. 2020, 98, 1255–1267. [Google Scholar] [CrossRef]
- Koscianska, E.; Starega-Roslan, J.; Krzyzosiak, W.J. The Role of Dicer Protein Partners in the Processing of MicroRNA Precursors. PLoS ONE 2011, 6, e28548. [Google Scholar] [CrossRef] [Green Version]
- Sha, L.-Y.; Zhang, Y.; Wang, W.; Sui, X.; Liu, S.-K.; Wang, T.; Zhang, H. MiR-18a upregulation decreases Dicer expression and confers paclitaxel resistance in triple negative breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2201–2208. [Google Scholar]
- Li, C.; Chen, L.; Song, W.; Peng, B.; Zhu, J.; Fang, L. DICER activates autophagy and promotes cisplatin resistance in non-small cell lung cancer by binding with let-7i-5p. Acta Histochem. 2021, 123, 151788. [Google Scholar] [CrossRef]
- Fan, Y.-X.; Dai, Y.-Z.; Wang, X.-L.; Ren, Y.-Q.; Han, J.-J.; Zhang, H. MiR-18a upregulation enhances autophagy in triple negative cancer cells via inhibiting mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2194–2200. [Google Scholar] [PubMed]
- Tanida, S.; Mizoshita, T.; Ozeki, K.; Tsukamoto, H.; Kamiya, T.; Kataoka, H.; Sakamuro, D.; Joh, T. Mechanisms of Cisplatin-Induced Apoptosis and of Cisplatin Sensitivity: Potential of BIN1 to Act as a Potent Predictor of Cisplatin Sensitivity in Gastric Cancer Treatment. Int. J. Surg. Oncol. 2012, 2012, 862879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabik, C.A.; Dolan, M.E. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007, 33, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Basourakos, S.P.; Li, L.; Aparicio, A.M.; Corn, P.G.; Kim, J.; Thompson, T.C. Combination Platinum-based and DNA Damage Response-targeting Cancer Therapy: Evolution and Future Directions. Curr. Med. Chem. 2017, 24, 1586–1606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Kang, Y.; Chen, L.; Wang, H.; Liu, J.; Zeng, S.; Yu, L. The Drug-Resistance Mechanisms of Five Platinum-Based Anticancer Agents. Front. Pharmacol. 2020, 11, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Gu, C.; Zhong, D.; Shi, L.; Kong, Y.; Zhou, Z.; Liu, S. Induction of autophagy counteracts the anticancer effect of cisplatin in human esophageal cancer cells with acquired drug resistance. Cancer Lett. 2014, 355, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, Z.; Zhang, Y.; Chen, X.; Guo, S.; Lei, Y.; Xu, Y.; Ji, C.; Bi, Z.; Wang, K. HMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biol. Ther. 2015, 16, 511–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Cui, J.; Yang, X.-M.; Liu, Z.-Y.; Song, F.; Li, L.; Jiang, J.-L.; Chen, Z.-N. Cytoplasmic fragment of CD147 generated by regulated intramembrane proteolysis contributes to HCC by promoting autophagy. Cell Death Dis. 2017, 8, e2925. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Wang, W. Knockdown of galectin-1 facilitated cisplatin sensitivity by inhibiting autophagy in neuroblastoma cells. Chem. Interactions 2019, 297, 50–56. [Google Scholar] [CrossRef]
- Fukuda, T.; Oda, K.; Wada-Hiraike, O.; Sone, K.; Inaba, K.; Ikeda, Y.; Miyasaka, A.; Kashiyama, T.; Tanikawa, M.; Arimoto, T.; et al. The anti-malarial chloroquine suppresses proliferation and overcomes cisplatin resistance of endometrial cancer cells via autophagy inhibition. Gynecol. Oncol. 2015, 137, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Xu, Y.; Su, J.; Yu, H.; Kang, J.; Li, H.; Li, X.; Xie, Q.; Yu, C.; Sun, L.; et al. Autophagic flux promotes cisplatin resistance in human ovarian carcinoma cells through ATP-mediated lysosomal function. Int. J. Oncol. 2015, 47, 1890–1900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, A.A.; Thömmes, J. Recent advances in large-scale production of monoclonal antibodies and related proteins. Trends Biotechnol. 2010, 28, 253–261. [Google Scholar] [CrossRef]
- Cimino-Mathews, A.; Foote, J.B.; Emens, L.A. Immune targeting in breast cancer. Oncology 2015, 29, 375–385. [Google Scholar] [PubMed]
- Wang, B.; Rouzier, R.; Albarracin, C.T.; Şahin, A.; Wagner, P.; Yang, Y.; Smith, T.L.; Meric-Bernstam, F.; Marcelo, A.C.; Hortobagyi, G.N.; et al. Expression of sigma 1 receptor in human breast cancer. Breast Cancer Res. Treat. 2004, 87, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.M.; Thomas, J.D.; Haas, D.A.; Longen, C.G.; Oyer, H.M.; Tong, J.Y.; Kim, F.J. Small-Molecule Sigma1 Modulator Induces Autophagic Degradation of PD-L1. Mol. Cancer Res. 2018, 16, 243–255. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Chen, Y. Autophagy controls programmed death-ligand 1 expression on cancer cells (Review). Biomed. Rep. 2021, 15, 84. [Google Scholar] [CrossRef]
- Wang, H.; Bloom, O.; Zhang, M.; Vishnubhakat, J.M.; Ombrellino, M.; Che, J.; Frazier, A.; Yang, H.; Ivanova, S.; Borovikova, L.; et al. HMG-1 as a Late Mediator of Endotoxin Lethality in Mice. Science 1999, 285, 248–251. [Google Scholar] [CrossRef]
- Pullerits, R.; Jonsson, I.-M.; Verdrengh, M.; Bokarewa, M.; Andersson, U.; Erlandsson-Harris, H.; Tarkowski, A. High mobility group box chromosomal protein 1, a DNA binding cytokine, induces arthritis. Arthritis Rheum. 2003, 48, 1693–1700. [Google Scholar] [CrossRef]
- Popovic, K.; Ek, M.; Espinosa, A.; Padyukov, L.; Harris, H.E.; Wahren-Herlenius, M.; Nyberg, F. Increased expression of the novel proinflammatory cytokine high mobility group box chromosomal protein 1 in skin lesions of patients with lupus erythematosus. Arthritis Rheum. 2005, 52, 3639–3645. [Google Scholar] [CrossRef]
- Zong, M.; Bruton, J.D.; Grundtman, C.; Yang, H.; Li, J.H.; Alexanderson, H.; Palmblad, K.; Andersson, U.; Harris, H.E.; Lundberg, I.E.; et al. TLR4 as receptor for HMGB1 induced muscle dysfunction in myositis. Ann. Rheum. Dis. 2013, 72, 1390–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.S.; Svetkauskaite, D.; He, Q.; Kim, J.Y.; Strassheim, D.; Ishizaka, A.; Abraham, E. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J. Biol. Chem. 2004, 279, 7370–7377. [Google Scholar] [CrossRef] [Green Version]
- Andersson, U.; Wang, H.; Palmblad, K.; Aveberger, A.-C.; Bloom, O.; Erlandsson-Harris, H.; Janson, A.; Kokkola, R.; Zhang, M.; Yang, H.; et al. High Mobility Group 1 Protein (Hmg-1) Stimulates Proinflammatory Cytokine Synthesis in Human Monocytes. J. Exp. Med. 2000, 192, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, Q.; Yang, H.; Tracey, K.J.; Bustin, M.; Oppenheim, J.J. High mobility group box-1 protein induces the migration and activation of human dendritic cells and acts as an alarmin. J. Leukoc. Biol. 2007, 81, 59–66. [Google Scholar] [CrossRef]
- Hubert, P.; Roncarati, P.; Demoulin, S.; Pilard, C.; Ancion, M.; Reynders, C.; Lerho, T.; Bruyere, D.; Lebeau, A.; Radermecker, C.; et al. Extracellular HMGB1 blockade inhibits tumor growth through profoundly remodeling immune microenvironment and enhances checkpoint inhibitor-based immunotherapy. J. Immunother. Cancer 2021, 9, e001966. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Venida, A.; Yano, J.; Biancur, D.E.; Kakiuchi, M.; Gupta, S.; Sohn, A.S.W.; Mukhopadhyay, S.; Lin, E.Y.; Parker, S.J.; et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 2020, 581, 100–105. [Google Scholar] [CrossRef]
- Loi, M.; Müller, A.; Steinbach, K.; Niven, J.; Barreira da Silva, R.; Paul, P.; Ligeon, L.-A.; Caruso, A.; Albrecht, R.A.; Becker, A.C.; et al. Macroautophagy Proteins Control MHC Class I Levels on Dendritic Cells and Shape Anti-viral CD8(+) T Cell Responses. Cell Rep. 2016, 15, 1076–1087. [Google Scholar] [CrossRef] [Green Version]
- DeVorkin, L.; Pavey, N.; Carleton, G.; Comber, A.; Ho, C.; Lim, J.; McNamara, E.; Huang, H.; Kim, P.; Zacharias, L.G.; et al. Autophagy Regulation of Metabolism Is Required for CD8(+) T Cell Anti-tumor Immunity. Cell Rep. 2019, 27, 502–513.e5. [Google Scholar] [CrossRef] [Green Version]
- Schrezenmeier, E.; Dörner, T. Mechanisms of action of hydroxychloroquine and chloroquine: Implications for rheumatology. Nat. Rev. Rheumatol. 2020, 16, 155–166. [Google Scholar] [CrossRef]
- Cook, K.L.; Wärri, A.; Soto-Pantoja, D.R.; Clarke, P.A.; Cruz, M.I.; Zwart, A.; Clarke, R. Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin. Cancer Res. 2014, 20, 3222–3232. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-O.; Mustafa, A.; Hudes, G.R.; Kruger, W.D. Hydroxychloroquine Destabilizes Phospho-S6 in Human Renal Carcinoma Cells. PLoS ONE 2015, 10, e0131464. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.-T.; Yu, X.-X.; Yan, L.-J.; Xiao, H.-T. Research progress of hydroxychloroquine and autophagy inhibitors on cancer. Cancer Chemother. Pharmacol. 2017, 79, 287–294. [Google Scholar] [CrossRef]
- Leung, L.-S.B.; Neal, J.W.; Wakelee, H.A.; Sequist, L.V.; Marmor, M.F. Rapid Onset of Retinal Toxicity from High-Dose Hydroxychloroquine Given for Cancer Therapy. Am. J. Ophthalmol. 2015, 160, 799–805.e1. [Google Scholar] [CrossRef]
- Carmichael, S.J.; Charles, B.; Tett, S.E. Population Pharmacokinetics of Hydroxychloroquine in Patients with Rheumatoid Arthritis. Ther. Drug Monit. 2003, 25, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Espina, V.A.; Liotta, L.; Rassulova, S.; Gallimore, H.; Grant-Wisdom, T.; Menezes, G.; Nayer, H.; Edmiston, K. Abstract CT140: PINC trial: Preventing invasive breast neoplasia with chloroquine. Cancer Res. 2017, 77, CT140. [Google Scholar] [CrossRef]
- Qiu, X.; Mei, J.; Yin, J.; Wang, H.; Wang, J.; Xie, M. Correlation analysis between expression of PCNA, Ki-67 and COX-2 and X-ray features in mammography in breast cancer. Oncol. Lett. 2017, 14, 2912–2918. [Google Scholar] [CrossRef] [Green Version]
- Anand, K.; Niravath, P.; Patel, T.; Ensor, J.; Rodriguez, A.; Boone, T.; Wong, S.T.; Chang, J.C. A Phase II Study of the Efficacy and Safety of Chloroquine in Combination with Taxanes in the Treatment of Patients With Advanced or Metastatic Anthracycline-refractory Breast Cancer. Clin. Breast Cancer 2021, 21, 199–204. [Google Scholar] [CrossRef]
- Pareek, A.; Suthar, M.; Rathore, G.S.; Bansal, V. Feverfew (Tanacetum parthenium L.): A systematic review. Pharmacogn. Rev. 2011, 5, 103–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Ong, C.-N.; Shen, H.-M. Critical roles of intracellular thiols and calcium in parthenolide-induced apoptosis in human colorectal cancer cells. Cancer Lett. 2004, 208, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Liu, L.; Lee, S.-O.; Kim, Y.-T.; You, K.-R.; Kim, D.-G. Susceptibility of Cholangiocarcinoma Cells to Parthenolide-Induced Apoptosis. Cancer Res. 2005, 65, 6312–6320. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-W.; Cai, M.-X.; Xin, Y.; Wu, Q.-S.; Ma, J.; Yang, P.; Xie, H.-Y.; Huang, D.-S. Parthenolide induces proliferation inhibition and apoptosis of pancreatic cancer cells in vitro. J. Exp. Clin. Cancer Res. 2010, 29, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, J.; You, K.R.; Lee, S.Y.; Song, C.H.; Kim, D.G. Oxidative stress-mediated apoptosis. The anticancer effect of the sesquiterpene lactone parthenolide. J. Biol. Chem. 2002, 277, 38954–38964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzman, M.L.; Rossi, R.M.; Karnischky, L.; Li, X.; Peterson, D.R.; Howard, D.S.; Jordan, C.T. The sesquiterpene lactone parthenolide induces apoptosis of human acute myelogenous leukemia stem and progenitor cells. Blood 2005, 105, 4163–4169. [Google Scholar] [CrossRef] [PubMed]
- Yip-Schneider, M.T.; Nakshatri, H.; Sweeney, C.J.; Marshall, M.S.; Wiebke, E.A.; Schmidt, C.M. Parthenolide and sulindac cooperate to mediate growth suppression and inhibit the nuclear factor-κB pathway in pancreatic carcinoma cells. Mol. Cancer Ther. 2005, 4, 587–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Adachi, M.; Kawamura, R.; Sakamoto, H.; Hayashi, T.; Ishida, T.; Imai, K.; Shinomura, Y. Parthenolide-induced apoptosis in multiple myeloma cells involves reactive oxygen species generation and cell sensitivity depends on catalase activity. Apoptosis 2006, 11, 2225–2235. [Google Scholar] [CrossRef]
- Zunino, S.J.; Ducore, J.M.; Storms, D.H. Parthenolide induces significant apoptosis and production of reactive oxygen species in high-risk pre-B leukemia cells. Cancer Lett. 2007, 254, 119–127. [Google Scholar] [CrossRef]
- Lu, C.; Wang, W.; Jia, Y.; Liu, X.; Tong, Z.; Li, B. Inhibition of AMPK/Autophagy Potentiates Parthenolide-Induced Apoptosis in Human Breast Cancer Cells. J. Cell. Biochem. 2014, 115, 1458–1466. [Google Scholar] [CrossRef]
- Luo, L.; Wang, J.N.; Kong, L.D.; Jiang, Q.G.; Tan, R.X. Antidepressant effects of Banxia Houpu decoction, a traditional Chinese medicinal empirical formula. J. Ethnopharmacol. 2000, 73, 277–281. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Lee, Y.M.; Lee, C.-K.; Jung, J.K.; Han, S.B.; Hong, J.T. Therapeutic applications of compounds in the Magnolia family. Pharmacol. Ther. 2011, 130, 157–176. [Google Scholar] [CrossRef]
- Liu, H.; Zang, C.; Emde, A.; Planas-Silva, M.D.; Rosche, M.; Kühnl, A.; Schulz, C.-O.; Elstner, E.; Possinger, K.; Eucker, J. Anti-tumor effect of honokiol alone and in combination with other anti-cancer agents in breast cancer. Eur. J. Pharmacol. 2008, 591, 43–51. [Google Scholar] [CrossRef]
- Wolf, I.; O’Kelly, J.; Wakimoto, N.; Nguyen, A.; Amblard, F.; Karlan, B.Y.; Arbiser, J.L.; Koeffler, H.P. Honokiol, a natural biphenyl, inhibits in vitro and in vivo growth of breast cancer through induction of apoptosis and cell cycle arrest. Int. J. Oncol. 2007, 30, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Muniraj, N.; Siddharth, S.; Shriver, M.; Nagalingam, A.; Parida, S.; Woo, J.; Elsey, J.; Gabrielson, K.; Gabrielson, E.; Arbiser, J.L.; et al. Induction of STK11-dependent cytoprotective autophagy in breast cancer cells upon honokiol treatment. Cell Death Discov. 2020, 6, 81. [Google Scholar] [CrossRef]
- Singh, N.; Bhalla, M.; De Jager, P.; Gilca, M. An Overview on Ashwagandha: A Rasayana (Rejuvenator) of Ayurveda. Afr. J. Tradit. Complement. Altern. Med. 2011, 8, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Vanden Berghe, W.; Sabbe, L.; Kaileh, M.; Haegeman, G.; Heyninck, K. Molecular insight in the multifunctional activities of Withaferin A. Biochem. Pharmacol. 2012, 84, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Vyas, A.R.; Singh, S.V. Molecular Targets and Mechanisms of Cancer Prevention and Treatment by Withaferin A, A Naturally Occurring Steroidal Lactone. AAPS J. 2014, 16, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, K.; De, S.; Mukherjee, S.; Das, S.; Ghosh, A.N.; Sengupta, S. Withaferin A induced impaired autophagy and unfolded protein response in human breast cancer cell-lines MCF-7 and MDA-MB-231. Toxicol. Vitr. 2017, 44, 330–338. [Google Scholar] [CrossRef]
- Muniraj, N.; Siddharth, S.; Nagalingam, A.; Walker, A.; Woo, J.; Győrffy, B.; Gabrielson, E.; Saxena, N.K.; Sharma, D. Withaferin A inhibits lysosomal activity to block autophagic flux and induces apoptosis via energetic impairment in breast cancer cells. Carcinogenesis 2019, 40, 1110–1120. [Google Scholar] [CrossRef]
- Button, R.W.; Roberts, S.L.; Willis, T.L.; Hanemann, C.O.; Luo, S. Accumulation of autophagosomes confers cytotoxicity. J. Biol. Chem. 2017, 292, 13599–13614. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Wang, J.; Liu, X.; Zhang, L.; Yi, G.; Li, C.; He, X.; Wang, P.; Jiang, H. Toosendanin Inhibits Hepatocellular Carcinoma Cells by Inducing Mitochondria-dependent Apoptosis. Planta Med. 2010, 76, 1447–1453. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.-L.; Li, M.-F. Biological effects of toosendanin, a triterpenoid extracted from Chinese traditional medicine. Prog. Neurobiol. 2007, 82, 1–10. [Google Scholar] [CrossRef]
- Kai, W.; Yating, S.; Lin, M.; Kaiyong, Y.; Baojin, H.; Wu, Y.; Fangzhou, Y.; Yan, C. Natural product toosendanin reverses the resistance of human breast cancer cells to adriamycin as a novel PI3K inhibitor. Biochem. Pharmacol. 2018, 152, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Dong, Y.; Chen, X.; Tan, C.S.H.; Li, M.; Miao, K.; Lu, J.-H. Toosendanin, a late-stage autophagy inhibitor, sensitizes triple-negative breast cancer to irinotecan chemotherapy. Chin. Med. 2022, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Nuta, G.C.; Gilad, Y.; Goldberg, N.; Meril, S.; Bahlsen, M.; Carvalho, S.; Kozer, N.; Barr, H.; Fridmann Sirkis, Y.; Hercík, K.; et al. Identifying a selective inhibitor of autophagy that targets ATG12-ATG3 protein-protein interaction. Autophagy 2023, 1–14. [Google Scholar] [CrossRef]
- Cui, J.; Ogasawara, Y.; Kurata, I.; Matoba, K.; Fujioka, Y.; Noda, N.N.; Shibasaki, M.; Watanabe, T. Targeting the ATG5-ATG16L1 Protein–Protein Interaction with a Hydrocarbon-Stapled Peptide Derived from ATG16L1 for Autophagy Inhibition. J. Am. Chem. Soc. 2022, 144, 17671–17679. [Google Scholar] [CrossRef]
- Sun, Q.; Hou, X.; Yang, J.; Zhang, M.; Yang, Y.; Liu, Y.; Shen, W.; Yin, D. Heparin-Coated Photosensitive Metal–Organic Frameworks as Drug Delivery Nanoplatforms of Autophagy Inhibitors for Sensitized Photodynamic Therapy against Breast Cancer. ACS Appl. Mater. Interfaces 2021, 13, 55577–55590. [Google Scholar] [CrossRef]
- Qian, R.; Cao, G.; Su, W.; Zhang, J.; Jiang, Y.; Song, H.; Jia, F.; Wang, H. Enhanced Sensitivity of Tumor Cells to Autophagy Inhibitors Using Fasting-Mimicking Diet and Targeted Lysosomal Delivery Nanoplatform. Nano Lett. 2022, 22, 9154–9162. [Google Scholar] [CrossRef]





| Phase | Clinical Trial | Treatment | Identifier |
|---|---|---|---|
| Unknown or Terminated or Withdrawn or Completed | |||
| 2 | Autophagy Inhibition Using Hydrochloroquine in Breast Cancer Patients: a Pilot Study | Hydroxychloroquine | NCT01292408 (Unknown status) 2012 |
| 1/2 | Phase I/II Study of Ixabepilone in Combination With the Autophagy Inhibitor Hydroxychloroquine for the Treatment of Patients with Metastatic Breast Cancer | Ixabepilone and Hydroxychloroquine | NCT00765765 (Terminated) 2013 |
| 2 | Phase Ib/II Study of Hydroxychloroquine in Metastatic ER-Positive Breast Cancer Progressing on Hormonal Therapy | Hydroxychloroquine in combination with hormonal therapy | NCT02414776 (Terminated) 2015 |
| 1/2 | Preventing Invasive Breast Neoplasia with Chloroquine (PINC) Trial | High or low dose of Chloroquine | NCT01023477 (Completed) 2016 |
| 2 | A Phase 2 Randomized, Double-blind, Window of Opportunity Trial Evaluating Trial Clinical and Correlative Effects of Chloroquine as a Novel Therapeutic Strategy in Breast Cancer | Chloroquine and placebo | NCT02333890 (Unknown) 2016 |
| 1/2 | A Phase Ib/II Trial of Gedatolisib, Hydroxychloroquine or the Combination for Prevention of Recurrent Breast Cancer (“GLACIER”) | Gedatolisib or Hydroxychloroquine alone or in combination | NCT03400254 (Withdrawn) 2020 |
| 2 | Phase II Study of The Efficacy and Safety of Chloroquine (C) in CombinAtion With Taxane or taxane-like (T) Chemo Agents in The Treatment of Patients With Advanced or Metastatic Breast Cancer Who Have Failed Anthracycline Chemo Base Therapy | Chloroquine with taxane or taxane-like chemotherapy | NCT01446016 (Completed) 2022 |
| Recruiting or Active | |||
| 1 | Hydroxychloroquine (HCQ) in Combination with Abemaciclib and Endocrine Therapy in HR+/Her2- Advanced Breast Cancer After a Lead in Dose Escalation Cohort of HCQ and Abemaciclib in Advanced Solid Tumors | Abemaciclib with alternative dose of HCQ or Abemaciclib with both HCQ and endocrine therapy | NCT04316169 (Recruiting) 2028 |
| 2 | A Phase II Pilot Trial of ABmacocliB or Abemaciclin and HydroxYchloroquine to Target Minimal Residual Disease in Brease Cancer Patients | Abemaciclib with or without Hydroxychloroquine | NCT04523857 (Recruiting) 2028 |
| 2 | A Phase II Trial of Avelumab or Hydrochloroquine With or Without Palbociclib to Eliminate Dormant Breast Cancer (PALAVY) | HCQ alone, or Avelumab alone, or Avelumab with Palbociclib, or HCQ with Palbociclib | NCT04841148 (Recruiting) 2028 |
| 1/2 | Phase I/II Safety and Efficacy Study of Autophagy Inhibition With Hydroxychloroquine to Augment the Antiproliferative and Biological Effects of Pre-Operative Palbociclib Plus Letrozole for Estrogen Receptor-Positive and HER2-Negative Breast Cancer | Hydroxycholoroquine, Letrozole, and Palbociclib | NCT03774472 (Active) 2024 |
| 2 | CLEVER Pilot Trial: A Phase II Pilot Trial of HydroxyChLoroquine, EVErolimus or the Combination for Prevention of Recurrent Breast Cancer | Hydroxychloroquine alone, or Everolimus alone, or the combination of hydroxychloroquine and Everolimus | NCT03032406 (Recruiting) 2025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Sharma, D. Autophagy and Breast Cancer: Connected in Growth, Progression, and Therapy. Cells 2023, 12, 1156. https://doi.org/10.3390/cells12081156
Wu Q, Sharma D. Autophagy and Breast Cancer: Connected in Growth, Progression, and Therapy. Cells. 2023; 12(8):1156. https://doi.org/10.3390/cells12081156
Chicago/Turabian StyleWu, Qitong, and Dipali Sharma. 2023. "Autophagy and Breast Cancer: Connected in Growth, Progression, and Therapy" Cells 12, no. 8: 1156. https://doi.org/10.3390/cells12081156
APA StyleWu, Q., & Sharma, D. (2023). Autophagy and Breast Cancer: Connected in Growth, Progression, and Therapy. Cells, 12(8), 1156. https://doi.org/10.3390/cells12081156