Multiple and Consecutive Genome Editing Using i-GONAD and Breeding Enrichment Facilitates the Production of Genetically Modified Mice
Abstract
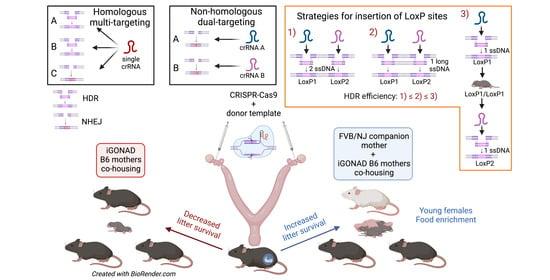
:1. Introduction
2. Material and Methods
2.1. Mice
2.2. CRISPR Reagents and Design
2.3. Hormone Treatment and Natural Mating
2.4. i-GONAD
2.5. Food Enrichment, Fostering, and Co-Rearing
2.6. Genetic Characterization of Founders and Data Analysis
3. Results
3.1. i-GONAD Reduces Litter Size
3.2. Food Enrichment, However, Not Natural Mating, Increases Pregnancy Rate in B6 Females
3.3. Co-Housing with Synchronized Good Mothers but Not Non-Survival Cesarean and Fostering Increases the Survival of i-GONAD B6 Pups
3.4. Using i-GONAD to Produce Gene Knockouts, Point Mutations, and Promoter Modifications
3.5. Using i-GONAD to Produce Conditional Knockout Mice
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brinster, R.L.; Chen, H.Y.; Warren, R.; Sarthy, A.; Palmiter, R.D. Regulation of metallothionein—Thymidine kinase fusion plasmids injected into mouse eggs. Nature 1982, 296, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Costantini, F.; Lacy, E. Introduction of a rabbit beta-globin gene into the mouse germ line. Nature 1981, 294, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.L.; Kelly, E.M.; Hunsicker, P.R.; Bangham, J.W.; Maddux, S.C.; Phipps, E.L. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc. Natl. Acad. Sci. USA 1979, 76, 5818–5819. [Google Scholar] [CrossRef]
- Gurumurthy, C.B.; Sato, M.; Nakamura, A.; Inui, M.; Kawano, N.; Islam, M.A.; Ogiwara, S.; Takabayashi, S.; Matsuyama, M.; Nakagawa, S.; et al. Creation of CRISPR-based germline-genome-engineered mice without ex vivo handling of zygotes by i-GONAD. Nat. Protoc. 2019, 14, 2452–2482. [Google Scholar] [CrossRef]
- Takahashi, G.; Gurumurthy, C.B.; Wada, K.; Miura, H.; Sato, M.; Ohtsuka, M. GONAD: Genome-editing via Oviductal Nucleic Acids Delivery system: A novel microinjection independent genome engineering method in mice. Sci. Rep. 2015, 5, 11406. [Google Scholar] [CrossRef]
- Hogan, B. Manipulating the Mouse Embryo: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1994; p. 497. [Google Scholar]
- Auerbach, A.B.; Norinsky, R.; Ho, W.; Losos, K.; Guo, Q.; Chatterjee, S.; Joyner, A.L. Strain-dependent differences in the efficiency of transgenic mouse production. Transgenic Res. 2003, 12, 59–69. [Google Scholar] [CrossRef]
- Concordet, J.P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- McLean, A.C.; Valenzuela, N.; Fai, S.; Bennett, S.A. Performing vaginal lavage, crystal violet staining, and vaginal cytological evaluation for mouse estrous cycle staging identification. J. Vis. Exp. 2012, 67, e4389. [Google Scholar] [CrossRef]
- Davis, M.W.; Jorgensen, E.M. ApE, A Plasmid Editor: A Freely Available DNA Manipulation and Visualization Program. Front. Bioinform. 2022, 2, 818619. [Google Scholar] [CrossRef] [PubMed]
- Melo-Silva, C.R.; Alves-Peixoto, P.; Heath, N.; Tang, L.; Montoya, B.; Knudson, C.J.; Stotesbury, C.; Ferez, M.; Wong, E.; Sigal, L.J. Resistance to lethal ectromelia virus infection requires Type I interferon receptor in natural killer cells and monocytes but not in adaptive immune or parenchymal cells. PLoS Pathog. 2021, 17, e1009593. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-P.; Li, X.-L.; Neises, A.; Chen, W.; Hu, L.-P.; Ji, G.-Z.; Yu, J.-Y.; Xu, J.; Yuan, W.-P.; Cheng, T.; et al. Different Effects of sgRNA Length on CRISPR-mediated Gene Knockout Efficiency. Sci. Rep. 2016, 6, 28566. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Zuñiga, J.; Edison, E.; Palla, S.; Dong, W.; Parker-Thornburg, J. Superovulation strategies for 6 commonly used mouse strains. J. Am. Assoc. Lab. Anim. Sci. 2011, 50, 471–478. [Google Scholar] [PubMed]
- Wilson, E.D.; Edwards, R.G. Parturition and increased litter size in mice after superovulation. J. Reprod Fertil. 1963, 5, 179–186. [Google Scholar] [CrossRef]
- Taketo, M.; Schroeder, A.C.; Mobraaten, L.E.; Gunning, K.B.; Hanten, G.; Fox, R.R.; Roderick, T.H.; Stewart, C.L.; Lilly, F.; Hansen, C.T.; et al. FVB/N: An inbred mouse strain preferable for transgenic analyses. Proc. Natl. Acad. Sci. USA 1991, 88, 2065–2069. [Google Scholar] [CrossRef] [PubMed]
- Laboratory, R.B.J.M.; Green, L. Handbook on Genetically Standardized Jax Mice; Jackson Laboratory: Bar Harbor, ME, USA, 1964. [Google Scholar]
- Kobayashi, Y.; Aoshima, T.; Ito, R.; Shinmura, R.; Ohtsuka, M.; Akasaka, E.; Sato, M.; Takabayashi, S. Modification of i-GONAD Suitable for Production of Genome-Edited C57BL/6 Inbred Mouse Strain. Cells 2020, 9, 957. [Google Scholar] [CrossRef] [PubMed]
- Thanos, D.; Maniatis, T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 1995, 83, 1091–1100. [Google Scholar] [CrossRef]
- Chen, D.; Chao, D.L.; Rocha, L.; Kolar, M.; Nguyen Huu, V.A.; Krawczyk, M.; Dasyani, M.; Wang, T.; Jafari, M.; Jabari, M.; et al. The lipid elongation enzyme ELOVL2 is a molecular regulator of aging in the retina. Aging. Cell 2020, 19, e13100. [Google Scholar] [CrossRef]
- Zadravec, D.; Tvrdik, P.; Guillou, H.; Haslam, R.; Kobayashi, T.; Napier, J.A.; Capecchi, M.R.; Jacobsson, A. ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. J. Lipid Res. 2011, 52, 245–255. [Google Scholar] [CrossRef]
- Pauter, A.M.; Olsson, P.; Asadi, A.; Herslöf, B.; Csikasz, R.I.; Zadravec, D.; Jacobsson, A. Elovl2 ablation demonstrates that systemic DHA is endogenously produced and is essential for lipid homeostasis in mice. J. Lipid Res. 2014, 55, 718–728. [Google Scholar] [CrossRef]
- Gregersen, L.H.; Schueler, M.; Munschauer, M.; Mastrobuoni, G.; Chen, W.; Kempa, S.; Dieterich, C.; Landthaler, M. MOV10 Is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol. Cell 2014, 54, 573–585. [Google Scholar] [CrossRef]
- Nawaz, A.; Shilikbay, T.; Skariah, G.; Ceman, S. Unwinding the roles of RNA helicase MOV10. Wiley Interdiscip Rev. RNA 2022, 13, e1682. [Google Scholar] [CrossRef] [PubMed]
- Skariah, G.; Seimetz, J.; Norsworthy, M.; Lannom, M.C.; Kenny, P.J.; Elrakhawy, M.; Forsthoefel, C.; Drnevich, J.; Kalsotra, A.; Ceman, S. Mov10 suppresses retroelements and regulates neuronal development and function in the developing brain. BMC Biol. 2017, 15, 54. [Google Scholar] [CrossRef]
- Fu, K.; Tian, S.; Tan, H.; Wang, C.; Wang, H.; Wang, M.; Wang, Y.; Chen, Z.; Wang, Y.; Yue, Q.; et al. Biological and RNA regulatory function of MOV10 in mammalian germ cells. BMC Biol. 2019, 17, 39. [Google Scholar] [CrossRef] [PubMed]
- Skariah, G.; Perry, K.J.; Drnevich, J.; Henry, J.J.; Ceman, S. RNA helicase Mov10 is essential for gastrulation and central nervous system development. Dev. Dyn. 2018, 247, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Tanave, A.; Matsuyama, M.; Koide, T. Efficient genome editing in wild strains of mice using the i-GONAD method. Sci. Rep. 2022, 12, 13821. [Google Scholar] [CrossRef]
- Shang, R.; Zhang, H.; Bi, P. Generation of mouse conditional knockout alleles in one step using the i-GONAD method. Genome Res. 2021, 31, 121–130. [Google Scholar] [CrossRef]
- Gurumurthy, C.B.; O’Brien, A.R.; Quadros, R.M.; Adams, J., Jr.; Alcaide, P.; Ayabe, S.; Ballard, J.; Batra, S.K.; Beauchamp, M.C.; Becker, K.A.; et al. Reproducibility of CRISPR-Cas9 methods for generation of conditional mouse alleles: A multi-center evaluation. Genome Biol. 2019, 20, 171. [Google Scholar] [CrossRef]





| Transgenic Mouse | Edited Locus | Genome Modification | CRISPR Repair Template | Number of Founders | CRISPR Efficiency * |
|---|---|---|---|---|---|
| Gpr15DREmut1 (Van et al., unpublished) | Chr 12, Gpr15 gene promoter | Disruption of two AHR binding sites in Gpr15 gene locus by multiple nucleotide substitution | long ssDNA for specific nucleotide substitutions in AHR binding site in the Gpr15 gene promoter | 5 ** | 20% imprecise HDR 40% total |
| Gpr15DREΔ (Van et al., unpublished) | Chr 12, Gpr15 gene promoter | Full deletion around two AHR binding sites in Gpr15 locus | long ssDNA for specific nucleotide substitutions in AHR binding site in the Gpr15 gene promoter | 5 ** | 40% total |
| Elovl2C234W | Chr 13, Elovl2 gene | One nucleotide mutation for cysteine to tryptophan substitution in the amino acid residue 234 of ELOVL2 protein | ssDNA for specific SNP | 18 | 33% HDR 66% total |
| Adipor1fl/fl | Chr 1, Adipor1 gene | LoxP insertions flanking Adipor1 gene exon 2 | long ssDNA for linked dual LoxP insertions flanking target exon | 10 | 20% HDR |
| Mov10Δexon12−14 | Chr 3, Mov10 gene | Deletion of exons 12, 13, and 14 by NHEJ | long ssDNA for linked dual LoxP insertions flanking exons 12, 13, and 14 | 16 (2 i-GONAD procedures) | 18.7% HDR on only 1 side 25% flanking sequence excision 75% total |
| Occ1Δexon3 | Chr 10, Occ1 gene | Deletion of exon3 by NHEJ | 2 ssDNA for one-step independent dual LoxP insertions flanking exon 3 | 4 | 50% HDR on only 1 side 50% flanking sequence excision 75% total |
| Rgs10−/− | Chr 7, Rgs10 gene | STOP codon insertion | ssDNA for STOP codon insertion in ORF | 9 | 44% HDR 67% total |
| Rgs1−/− | Chr 1, Rgs1 gene | STOP codon insertion | ssDNA for STOP codon insertion in ORF | 8 | 25% HDR 37.5% total |
| Ifnb1ΔPRDII | Chr 4, Ifnb1 gene promoter | PRDII/NF-κβ binding site deletion in Ifnb1 gene promoter | ssDNA for specific nucleotide deletions in promoter | 1 | 100% total |
| Ifnb1−/− | Chr 4, Ifnb1 gene | NHEJ indel resulting in early STOP codon | ssDNA for STOP codon insertion in ORF | 3 ** | 33% HDR 100% total |
| Ifna4,b1−/− | Chr 4, Ifna4 and Ifnb1 genes | STOP codon insertion in Ifna4 gene and NHEJ indel resulting in early STOP codon in Ifnb1 gene | ssDNA for STOP codon insertion in ORF | 3 ** | 33% HDR 100% total |
| Ifna4−/− | Chr 4, Ifna4 gene | STOP codon insertion | ssDNA for STOP codon insertion in ORF | 8 | 62% HDR 100% total |
| Ifna1,5,7−/− | Chr 4; Ifna1, Ifna5 and Ifna7 genes | STOP codon insertion in Ifna1 and Ifna5 genes and NHEJ indel resulting in early STOP codon in Ifna7 gene | ssDNA for STOP codon insertion in ORF | 7 | 14% HDR 28% total |
| Ifna6-16−/− | Chr 4, Ifna locus | Large chromosome deletion by NHEJ resulting in deletion of Ifna16, Ifna2, Ifnab, Ifna7, Ifna11, Ifna6, and Klhl9 genes | 2 ssDNA for one-step independent dual LoxP insertions in intergenic sequences | 5 | 40% precise HDR only on 1 insertion site 20% imprecise HDR |
| IfnaIfna1-Ifne fl | Chr 4, Ifna locus | upstream LoxP insertion at intergenic sequence (between Ifnb1 and Ifna15 genes) and downstream insertion of LoxP at intergenic sequence (between Ifna1 and Ifne genes) | 2 ssDNA for one-step independent dual LoxP insertions in intergenic sequences | 7 | 14% HDR only on downstream LoxP insertion |
| Ifnafl/fl | Chr 4, Ifna locus | upstream LoxP insertion at intergenic sequence (between Ifnb1 and Ifna15 genes) on IfnaIfna1-Ifne fl founder offspring | ssDNA a single LoxP insertion in intergenic sequence | 9 | 56% HDR |
| Breeders (Male, Female) * | Male Genotype | Female Genotype | # Number of Pups |
|---|---|---|---|
| WT B6, i-GONAD founder | Elovl2+/+ | Elovl2−/− | 40 |
| i-GONAD founder, WT B6 | Elovl2−/− | Elovl2+/+ | 0 |
| WT B6, F1 het | Elovl2+/+ | Elovl2+/− | 6 |
| F1 het, WT B6 | Elovl2+/− | Elovl2+/+ | 0 |
| WT B6, F1 het | Elovl2+/+ | Elovl2+/− | 14 |
| WT B6, F1 het | Elovl2+/+ | Elovl2+/− | 31 |
| WT B6, F1 het | Elovl2+/+ | Elovl2+/− | 21 |
| F1 het, WT B6 | Elovl2+/− | Elovl2+/+ | 0 |
| F1 het, WT B6 | Elovl2+/− | Elovl2+/+ | 0 |
| F1 het, WT B6 | Elovl2+/− | Elovl2+/+ | 0 |
| F1 het, F1 het | Elovl2+/− | Elovl2+/− | 0 |
| F1 het, F1 het | Elovl2+/− | Elovl2+/− | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melo-Silva, C.R.; Knudson, C.J.; Tang, L.; Kafle, S.; Springer, L.E.; Choi, J.; Snyder, C.M.; Wang, Y.; Kim, S.V.; Sigal, L.J. Multiple and Consecutive Genome Editing Using i-GONAD and Breeding Enrichment Facilitates the Production of Genetically Modified Mice. Cells 2023, 12, 1343. https://doi.org/10.3390/cells12091343
Melo-Silva CR, Knudson CJ, Tang L, Kafle S, Springer LE, Choi J, Snyder CM, Wang Y, Kim SV, Sigal LJ. Multiple and Consecutive Genome Editing Using i-GONAD and Breeding Enrichment Facilitates the Production of Genetically Modified Mice. Cells. 2023; 12(9):1343. https://doi.org/10.3390/cells12091343
Chicago/Turabian StyleMelo-Silva, Carolina R., Cory J. Knudson, Lingjuan Tang, Samita Kafle, Lauren E. Springer, Jihae Choi, Christopher M. Snyder, Yajing Wang, Sangwon V. Kim, and Luis J. Sigal. 2023. "Multiple and Consecutive Genome Editing Using i-GONAD and Breeding Enrichment Facilitates the Production of Genetically Modified Mice" Cells 12, no. 9: 1343. https://doi.org/10.3390/cells12091343
APA StyleMelo-Silva, C. R., Knudson, C. J., Tang, L., Kafle, S., Springer, L. E., Choi, J., Snyder, C. M., Wang, Y., Kim, S. V., & Sigal, L. J. (2023). Multiple and Consecutive Genome Editing Using i-GONAD and Breeding Enrichment Facilitates the Production of Genetically Modified Mice. Cells, 12(9), 1343. https://doi.org/10.3390/cells12091343