Modelling of Neuronal Ceroid Lipofuscinosis Type 2 in Dictyostelium discoideum Suggests That Cytopathological Outcomes Result from Altered TOR Signalling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Creation of Constructs
2.2. Transformation and Cell Culture
2.3. Calculation of Gene Copy Number
2.4. Determination of Relative mRNA Expression Levels
2.5. Autofluorescence Experiments
2.6. Growth Experiments
2.7. Endocytosis Experiments
2.8. Phototaxis Experiments
2.9. Calculation of Cell Numbers in A Slug
2.10. Rapamycin Experiments
3. Results
3.1. Creation of Transformants with Decreased Levels of Tpp1 Expression
3.2. Dictyostelium Tpp1 Mutants Display Autofluorescence
3.3. Tpp1 is Required for Normal Development, but Not for Phototaxis
3.4. Tpp1 Knockdown Causes Slow Growth but Elevated Rates of Endocytosis
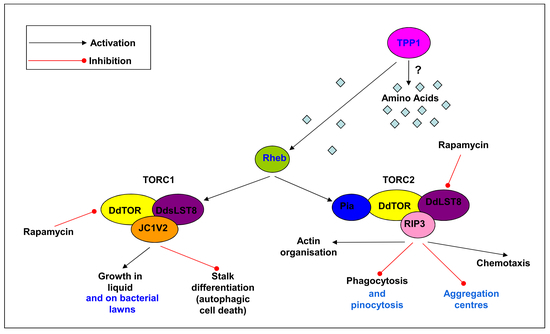
3.5. Pharmacological or Genetic Inhibition of TOR Signalling Phenocopies Loss of Tpp1
3.6. Overexpression of Rheb Rescues the Phenotypic Effects in the Tpp1 Mutants
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Cárcel-Trullols, J.; Kovács, A.D.; Pearce, D.A. Cell biology of the NCL proteins: What they do and don’t do. Biochim. Biophys. Acta 2015, 1852, 2242–2255. [Google Scholar] [CrossRef]
- Palmer, D.N.; Fearnley, I.M.; Walker, J.E.; Hall, N.A.; Lake, B.D.; Wolfe, L.S.; Haltia, M.; Martinus, R.D.; Jolly, R.D. Mitochondrial ATP synthase subunitc storage in the ceroid-lipofuscinoses (Batten disease). Am. J. Med. Genet. 1992, 42, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Kousi, M.; Lehesjoki, A.E.; Mole, S.E. Update of the mutation spectrum and clinical correlations of over 360 mutations in eight genes that underlie the neuronal ceroid lipofuscinoses. Hum. Mutat. 2012, 33, 42–63. [Google Scholar] [CrossRef]
- Jalanko, A.; Braulke, T. Neuronal ceroid lipofuscinoses. Biochim. Biophys. Acta 2009, 1793, 697–709. [Google Scholar] [CrossRef] [Green Version]
- Vines, D.; Warburton, M.J. Purification and characterisation of a tripeptidyl aminopeptidase I from rat spleen. Biochim. Biophys. Acta 1998, 1384, 233–242. [Google Scholar] [CrossRef]
- Ezaki, J.; Takeda-Ezaki, M.; Oda, K.; Kominami, E. Characterization of Endopeptidase Activity of Tripeptidyl Peptidase-I/CLN2 Protein Which Is Deficient in Classical Late Infantile Neuronal Ceroid Lipofuscinosis. Biochem. Biophys. Res. Commun. 2000, 268, 904–908. [Google Scholar] [CrossRef]
- Golabek, A.A.; Kida, E. Tripeptidyl-peptidase I in health and disease. Biol. Chem. 2006, 387, 1091–1099. [Google Scholar] [CrossRef]
- Bond, M.; Holthaus, S.M.; Tammen, I.; Tear, G.; Russell, C. Use of model organisms for the study of neuronal ceroid lipofuscinosis. Biochim. Biophys. Acta 2013, 1832, 1842–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faller, K.M.; Gutierrez-Quintana, R.; Mohammed, A.; Rahim, A.A.; Tuxworth, R.I.; Wager, K.; Bond, M. The neuronal ceroid lipofuscinoses: Opportunities from model systems. Biochim. Biophys. Acta 2015, 1852, 2267–2278. [Google Scholar] [CrossRef] [Green Version]
- Huber, R.J. Using the social amoeba Dictyostelium to study the functions of proteins linked to neuronal ceroid lipofuscinosis. J. Biomed. Sci. 2016, 23, 83. [Google Scholar] [CrossRef]
- Phillips, S.N.; Muzaffar, N.; Codlin, S.; Korey, C.A.; Taschner, P.E.; De Voer, G.; Mole, S.E.; Pearce, D.A. Characterizing pathogenic processes in Batten disease: Use of small eukaryotic model systems. Biochim. Biophys. Acta 2006, 1762, 906–919. [Google Scholar] [CrossRef] [Green Version]
- Annesley, S.J.; Fisher, P.R. Dictyostelium discoideum—a model for many reasons. Mol. Cell. Biochem. 2009, 329, 73–91. [Google Scholar] [CrossRef]
- Journet, A.; Chapel, A.; Jehan, S.; Adessi, C.; Freeze, H.; Klein, G.; Garin, J. Characterization of Dictyostelium discoideum cathepsin D. J. Cell Sci. 1999, 112, 3833–3843. [Google Scholar]
- Huber, R.J.; Mathavarajah, S. Cln5 is secreted and functions as a glycoside hydrolase in Dictyostelium. Cell. Signal. 2018, 42, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Huber, R.J.; Mathavarajah, S. Secretion and function of Cln5 during the early stages of Dictyostelium development. Biochim. Biophys. Acta Mol. Cell. Res. 2018, 1865, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Huber, R.J.; Myre, M.A.; Cotman, S.L. Loss of Cln3 Function in the Social Amoeba Dictyostelium discoideum Causes Pleiotropic Effects That Are Rescued by Human CLN3. PLoS ONE 2014, 9, 110544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, R.J. Loss of Cln3 impacts protein secretion in the social amoeba Dictyostelium. Cell. Signal. 2017, 35, 61–72. [Google Scholar] [CrossRef]
- Stumpf, M.; Müller, R.; Gaßen, B.; Wehrstedt, R.; Fey, P.; Karow, M.A.; Eichinger, L.; Glöckner, G.; Noegel, A.A. A tripeptidyl peptidase 1 is a binding partner of the Golgi pH regulator (GPHR) in Dictyostelium. Dis. Model. Mech. 2017, 10, 897–907. [Google Scholar] [CrossRef]
- Phillips, J.E.; Gomer, R.H. Partial genetic suppression of a loss-of-function mutant of the neuronal ceroid lipofuscinosis-associated protease TPP1 in Dictyostelium discoideum. Dis. Model. Mech. 2015, 8, 147–156. [Google Scholar] [CrossRef]
- Liao, X.-H.; Majithia, A.; Huang, X.; Kimmel, A.R. Growth control via TOR kinase signaling, an intracellular sensor of amino acid and energy availability, with crosstalk potential to proline metabolism. Amino Acids 2008, 35, 761–770. [Google Scholar] [CrossRef]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR Signaling in Growth and Metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [Green Version]
- Fey, P.; Compton, K.; Cox, E.C. Green fluorescent protein production in the cellular slime molds Polysphondylium pallidum and Dictyostelium discoideum. Gene 1995, 165, 127–130. [Google Scholar] [CrossRef]
- Nellen, W.; Silan, C.; Firtel, R.A. DNA-mediated transformation in Dictyostelium discoideum: Regulated expression of an actin gene fusion. Mol. Cell. Biol. 1984, 4, 2890–2898. [Google Scholar] [CrossRef]
- Wilczynska, Z.; Fisher, P.R. Analysis of a Complex Plasmid Insertion in a Phototaxis-Deficient Transformant of Dictyostelium discoideum Selected on a Micrococcus luteus Lawn. Plasmid 1994, 32, 182–194. [Google Scholar] [CrossRef]
- Barth, C.; Fraser, D.J.; Fisher, P.R. A rapid, small scale method for characterization of plasmid insertions in the Dictyostelium genome. Nucleic Acids Res. 1998, 26, 3317–3318. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Annesley, S.J.; Jasim, R.A.F.; Musco, V.J.; Sanislav, O.; Fisher, P.R. The Parkinson’s disease-associated protein DJ-1 plays a positive nonmitochondrial role in endocytosis in Dictyostelium cells. Dis. Model. Mech. 2017, 10, 1261–1271. [Google Scholar] [CrossRef]
- Laevsky, G.; Knecht, D.A.; Maselli, A. Kinetics of binding, uptake and degradation of live fluorescent (DsRed) bacteria by Dictyostelium discoideum. Microbiology 2002, 148, 413–420. [Google Scholar] [Green Version]
- Bokko, P.B.; Francione, L.; Bandala-Sanchez, E.; Ahmed, A.U.; Annesley, S.J.; Huang, X.; Khurana, T.; Kimmel, A.R.; Fisher, P.R. Diverse Cytopathologies in Mitochondrial Disease Are Caused by AMP-activated Protein Kinase Signaling. Mol. Biol. Cell 2007, 18, 1874–1886. [Google Scholar] [CrossRef] [Green Version]
- Annesley, S.J.; Fisher, P.R. Dictyostelium Slug Phototaxis. Methods Mol. Biol. 2009, 571, 67–76. [Google Scholar] [PubMed]
- Barth, C.; Fraser, D.J.; Fisher, P.R. Co-insertional Replication Is Responsible for Tandem Multimer Formation during Plasmid Integration into the Dictyostelium genome. Plasmid 1998, 39, 141–153. [Google Scholar] [CrossRef]
- Rot, G.; Parikh, A.; Curk, T.; Kuspa, A.; Shaulsky, G.; Zupan, B. DictyExpress: A Dictyostelium discoideum gene expression database with an explorative data analysis web-based interface. BMC Bioinform. 2009, 10, 265. [Google Scholar] [CrossRef]
- Smith, E.; Fisher, P.R.; Grant, W.N.; Williams, K.L. Sensory Behavior in Dictyostelium-Discoideum Slugs - Phototaxis and Thermotaxis Are Not Mediated by a Change in Slug Speed. J. Cell Sci. 1982, 54, 329–339. [Google Scholar]
- Schindler, J.; Sussman, M. Ammonia determines the choice of morphogenetic pathways in Dictyostelium discoideum. J. Mol. Biol. 1977, 116, 161–169. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Ali, S.M.; Sengupta, S.; Sheen, J.-H.; Hsu, P.P.; Bagley, A.F.; Markhard, A.L.; Sabatini, D.M. Prolonged Rapamycin Treatment Inhibits mTORC2 Assembly and Akt/PKB. Mol. Cell 2006, 22, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Iranfar, N.; Fuller, D.; Šášik, R.; Hwa, T.; Laub, M.; Loomis, W.F. Expression Patterns of Cell-type–specific Genes in Dictyostelium. Mol. Biol. Cell 2001, 12, 2590–2600. [Google Scholar] [CrossRef] [PubMed]
- Seehafer, S.S.; Pearce, D.A. You say lipofuscin, we say ceroid: Defining autofluorescent storage material. Neurobiol. Aging 2006, 27, 576–588. [Google Scholar] [CrossRef]
- Seehafer, S.S.; Pearce, D.A. Spectral properties and mechanisms that underlie autofluorescent accumulations in Batten disease. Biochem. Biophys. Res. Commun. 2009, 382, 247–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Voer, G.; van der Bent, P.; Rodrigues, A.J.; van Ommen, G.J.; Peters, D.J.; Taschner, P.E. Deletion of the Caenorhabditis elegans homologues of the CLN3 gene, involved in human juvenile neuronal ceroid lipofuscinosis, causes a mild progeric phenotype. J. Inherit. Metab. Dis. 2005, 28, 1065–1080. [Google Scholar] [CrossRef]
- Dimond, R.L.; Brenner, M.; Loomis, W.F., Jr. Mutations affecting N-acetylglucosaminidase in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 1973, 70, 3356–3360. [Google Scholar] [CrossRef]
- Maniak, M. Fusion and Fission Events in the Endocytic Pathway of Dictyostelium. Traffic 2003, 4, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Müller, I.; Šubert, N.; Otto, H.; Herbst, R.; Rühling, H.; Maniak, M.; Leippe, M. A Dictyostelium Mutant with Reduced Lysozyme Levels Compensates by Increased Phagocytic Activity. J. Biol. Chem. 2005, 280, 10435–10443. [Google Scholar] [CrossRef] [Green Version]
- Galardi-Castilla, M.; Pergolizzi, B.; Bloomfield, G.; Skelton, J.; Ivens, A.; Kay, R.R.; Bozzaro, S.; Sastre, L. SrfB, a member of the Serum Response Factor family of transcription factors, regulates starvation response and early development in Dictyostelium. Dev. Biol. 2008, 316, 260–274. [Google Scholar] [CrossRef] [Green Version]
- Phang, J.M.; Donald, S.P.; Pandhare, J.; Liu, Y. The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids 2008, 35, 681–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosel, D.; Khurana, T.; Majithia, A.; Huang, X.; Bhandari, R.; Kimmel, A.R. TOR complex 2 (TORC2) in Dictyostelium suppresses phagocytic nutrient capture independently of TORC1-mediated nutrient sensing. J. Cell Sci. 2012, 125, 37–48. [Google Scholar] [CrossRef]
- Urano, J.; Ellis, C.; Clark, G.J.; Tamanoi, F. Characterization of Rheb functions using yeast and mammalian systems. Methods Enzymol. 2001, 333, 217–231. [Google Scholar]
- Oh, W.J.; Jacinto, E. mTOR complex 2 signaling and functions. Cell Cycle 2011, 10, 2305–2316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loomis, W.F., Jr. Sensitivity of Dictyostelium discoideum to nucleic acid analogues. Exp. Cell 1971, 64, 484–486. [Google Scholar] [CrossRef]
- Watts, D.J.; Ashworth, J.M. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J. 1970, 119, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Bond, M.E.; Brown, R.; Rallis, C.; Bähler, J.; Mole, S.E. A central role for TOR signalling in a yeast model for juvenile CLN3 disease. Microb. Cell 2015, 2, 466–480. [Google Scholar] [CrossRef] [Green Version]











© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, P.K.; Sen, M.G.; Fisher, P.R.; Annesley, S.J. Modelling of Neuronal Ceroid Lipofuscinosis Type 2 in Dictyostelium discoideum Suggests That Cytopathological Outcomes Result from Altered TOR Signalling. Cells 2019, 8, 469. https://doi.org/10.3390/cells8050469
Smith PK, Sen MG, Fisher PR, Annesley SJ. Modelling of Neuronal Ceroid Lipofuscinosis Type 2 in Dictyostelium discoideum Suggests That Cytopathological Outcomes Result from Altered TOR Signalling. Cells. 2019; 8(5):469. https://doi.org/10.3390/cells8050469
Chicago/Turabian StyleSmith, Paige K., Melodi G. Sen, Paul R. Fisher, and Sarah J. Annesley. 2019. "Modelling of Neuronal Ceroid Lipofuscinosis Type 2 in Dictyostelium discoideum Suggests That Cytopathological Outcomes Result from Altered TOR Signalling" Cells 8, no. 5: 469. https://doi.org/10.3390/cells8050469
APA StyleSmith, P. K., Sen, M. G., Fisher, P. R., & Annesley, S. J. (2019). Modelling of Neuronal Ceroid Lipofuscinosis Type 2 in Dictyostelium discoideum Suggests That Cytopathological Outcomes Result from Altered TOR Signalling. Cells, 8(5), 469. https://doi.org/10.3390/cells8050469