Distinct Protein Expression Networks are Activated in Microglia Cells after Stimulation with IFN-γ and IL-4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Flow Cytometry Analysis
2.3. Real-Time PCR Analysis
2.4. Western Blot Analysis
2.5. Protein Extraction and Digestion
2.6. Mass Spectrometry Analysis, Database Searching, and Bioinformatics Analysis
3. Results
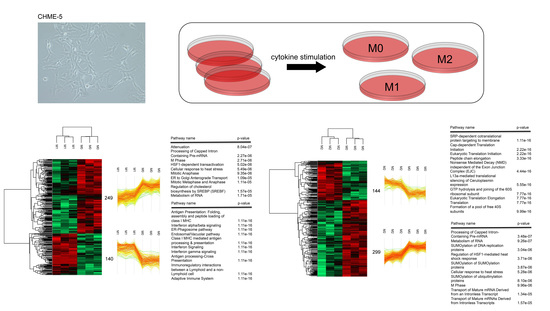
3.1. IFN-γ and IL-4 Treated CHME-5 Microglia Cells Presented Distinct M1- and M2-Like Phenotypes
3.2. Characterization of Proteomic Changes in Classically (M1) and Alternatively (M2) Activated CHME-5 Microglia Cells
3.3. Validation of Signaling and Metabolic Pathways Determined by Proteomics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lawson, L.J. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 1990, 39, 151–170. [Google Scholar] [CrossRef]
- Prinz, M.; Priller, J. Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat. Rev. Neurosci. 2014, 15, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Aguzzi, A.; Barres, B.A.; Bennett, M.L. Microglia: Scapegoat, saboteur, or something else? Science 2013, 339, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Tay, T.L.; Savage, J.C.; Hui, C.W.; Bisht, K.; Tremblay, M.È. Microglia across the lifespan: From origin to function in brain development, plasticity and cognition. J Physiol. 2017, 595, 1929–1945. [Google Scholar] [CrossRef] [PubMed]
- Voet, S.; Prinz, M.; Van Loo, G. Microglia in Central Nervous System Inflammation and Multiple Sclerosis Pathology. Trends Mol. Med. 2019, 25, 112–123. [Google Scholar] [CrossRef]
- Gautier, E.L.; Shay, T.; Miller, J.; Greter, M.; Jakubzick, C.; Ivanov, S.; Helft, J.; Chow, A.; Elpek, K.G.; Gordonov, S.; et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012, 13, 1118–1128. [Google Scholar] [CrossRef] [Green Version]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef]
- Greter, M.; Merad, M. Regulation of microglia development and homeostasis. Glia 2013, 61, 121–127. [Google Scholar] [CrossRef]
- Ajami, B.; Bennett, J.L.; Krieger, C.; Tetzlaff, W.; Rossi, F.M. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 2007, 10, 1538–1543. [Google Scholar] [CrossRef]
- Kettenmann, H.; Hanisch, U.K.; Noda, M.; Verkhratsky, A. Physiology of microglia. Physiol. Rev. 2011, 91, 461–553. [Google Scholar] [CrossRef]
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- Franco, R.; Fernández-Suárez, D. Alternatively activated microglia and macrophages in the central nervous system. Prog. Neurobiol. 2015, 131, 65–86. [Google Scholar] [CrossRef] [PubMed]
- Hanisch, U.K.; Kettenmann, H. Microglia: Active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 2007, 10, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Hickman, S.E.; Kingery, N.D.; Ohsumi, T.K.; Borowsky, M.L.; Wang, L.C.; Means, T.K.; El Khoury, J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013, 16, 1896–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rock, R.B.; Hu, S.; Deshpande, A.; Munir, S.; May, B.J.; Baker, C.A.; Peterson, P.K.; Kapur, V. Transcriptional response of human microglial cells to interferon-gamma. Genes Immun. 2005, 6, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.; Cusimano, M.; Zambito, M.; Finardi, A.; Capotondo, A.; Garcia-Manteiga, J.M.; Comi, G.; Furlan, R.; Martino, G.; Muzio, L. Interleukin 4 modulates microglia homeostasis and attenuates the early slowly progressive phase of amyotrophic lateral sclerosis. Cell Death Dis. 2018, 9, 250. [Google Scholar] [CrossRef] [PubMed]
- Janabi, N.; Peudenier, S.; Héron, B.; Ng, K.H.; Tardieu, M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci. Lett. 1995, 195, 105–108. [Google Scholar] [CrossRef]
- Garcia-Mesa, Y.; Jay, T.R.; Checkley, M.A.; Luttge, B.; Dobrowolski, C.; Valadkhan, S.; Landreth, G.E.; Karn, J.; Alvarez-Carbonell, D. Immortalization of primary microglia: A new platform to study HIV regulation in the central nervous system. J. Neurovirol. 2017, 23, 47–66. [Google Scholar] [CrossRef]
- Figueroa-Hall, L.K.; Anderson, M.B.; Das, S.; Stevens, C.; Davis, R.L. LPS-induced TLR4 neuroinflammatory signaling in CHME-5 microglial cells. Neuroimmunol. Neuroinflamm. 2017, 4, 219–231. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1118. [Google Scholar] [CrossRef]
- Vergara, D.; Stanca, E.; Guerra, F.; Priore, P.; Gaballo, A.; Franck, J.; Simeone, P.; Trerotola, M.; De Domenico, S.; Fournier, I.; et al. β-Catenin Knockdown Affects Mitochondrial Biogenesis and Lipid Metabolism in Breast Cancer Cells. Front. Physiol. 2017, 8, 544. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M. A polarizing question: Do M1 and M2 microglia exist? Nat. Neurosci. 2016, 19, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Togo, T.; Akiyama, H.; Kondo, H.; Ikeda, K.; Kato, M.; Iseki, E.; Kosaka, K. Expression of CD40 in the brain of Alzheimer’s disease and other neurological diseases. Brain Res. 2000, 885, 117–121. [Google Scholar] [CrossRef]
- Chen, K.; Huang, J.; Gong, W.; Zhang, L.; Yu, P.; Wang, J.M. CD40/CD40L dyad in the inflammatory and immune responses in the central nervous system. Cell. Mol. Immunol. 2006, 3, 163–169. [Google Scholar]
- Relloso, M.; Puig-Kröger, A.; Pello, O.M.; Rodríguez-Fernández, J.L.; De la Rosa, G.; Longo, N.; Navarro, J.; Muñoz-Fernández, M.A.; Sánchez-Mateos, P.; Corbí, A.L. DC-SIGN (CD209) expression is IL-4 dependent and is negatively regulated by IFN, TGF-beta, and anti-inflammatory agents. J. Immunol. 2002, 168, 2634–2643. [Google Scholar] [CrossRef] [PubMed]
- Peferoen, L.A.; Vogel, D.Y.; Ummenthum, K.; Breur, M.; Heijnen, P.D.; Gerritsen, W.H.; Peferoen-Baert, R.M.; van der Valk, P.; Dijkstra, C.D.; Amor, S. Activation status of human microglia is dependent on lesion formation stage and remyelination in multiple sclerosis. J. Neuropathol. Exp. Neurol. 2015, 74, 48–63. [Google Scholar] [CrossRef]
- Kaminska, B.; Mota, M.; Pizzi, M. Signal transduction and epigenetic mechanisms in the control of microglia activation during neuroinflammation. Biochim. Biophys. Acta 2016, 1862, 339–351. [Google Scholar]
- Wu, J.; Ocampo, A.; Belmonte, J.C.I. Cellular Metabolism and Induced Pluripotency. Cell 2016, 166, 1371–1385. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, L.M. The Role of Nuclear Factor κB in the Interferon Response. J. Interferon Cytokine Res. 2011, 31, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Bousoik, E.; Montazeri Aliabadi, H. “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway. Front. Oncol. 2018, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Stienstra, R. Specific and Complex Reprogramming of Cellular Metabolism in Myeloid Cells during Innate Immune Responses. Cell Metab. 2017, 26, 142–156. [Google Scholar] [CrossRef]
- Phan, A.T.; Goldrath, A.W.; Glass, C.K. Metabolic and Epigenetic Coordination of T Cell and Macrophage Immunity. Immunity 2017, 46, 714–729. [Google Scholar] [CrossRef] [Green Version]
- Biswas, S.K.; Mantovani, A. Orchestration of metabolism by macrophages. Cell Metab. 2012, 15, 432–437. [Google Scholar] [CrossRef]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef]
- Bell-Temin, H.; Culver-Cochran, A.E.; Chaput, D.; Carlson, C.M.; Kuehl, M.; Burkhardt, B.R.; Bickford, P.C.; Liu, B.; Stevens, S.M., Jr. Novel Molecular Insights into Classical and Alternative Activation States of Microglia as Revealed by Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)-based Proteomics. Mol. Cell Proteomics 2015, 14, 3173–3184. [Google Scholar] [CrossRef] [PubMed]
- Zissler, U.M.; Chaker, A.M.; Effner, R.; Ulrich, M.; Guerth, F.; Piontek, G.; Dietz, K.; Regn, M.; Knapp, B.; Theis, F.J.; et al. Interleukin-4 and interferon-γ orchestrate an epithelial polarization in the airways. Mucosal Immunol. 2016, 9, 917–926. [Google Scholar] [CrossRef]
- Boutej, H.; Rahimian, R.; Thammisetty, S.S.; Béland, L.C.; Lalancette-Hébert, M.; Kriz, J. Diverging mRNA and Protein Networks in Activated Microglia Reveal SRSF3 Suppresses Translation of Highly Upregulated Innate Immune Transcripts. Cell Rep. 2017, 21, 3220–3233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Donnell, L.A.; Henkins, K.M.; Kulkarni, A.; Matullo, C.M.; Balachandran, S.; Pattisapu, A.K.; Rall, G.F. Interferon gamma induces protective non-canonical signaling pathways in primary neurons. J. Neurochem. 2015, 135, 309–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zibara, K.; Zeidan, A.; Bjeije, H.; Kassem, N.; Badran, B.; El-Zein, N. ROS mediates interferon gamma induced phosphorylation of Src, through the Raf/ERK pathway, in MCF-7 human breast cancer cell line. J. Cell Commun. Signal. 2017, 11, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Meares, G.P.; Qin, H.; Liu, Y.; Holdbrooks, A.T.; Benveniste, E.N. AMP-activated protein kinase restricts IFN-γ signaling. J. Immunol. 2013, 190, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Gromovsky, A.D.; Schugar, R.C.; Brown, A.L.; Helsley, R.N.; Burrows, A.C.; Ferguson, D.; Zhang, R.; Sansbury, B.E.; Lee, R.G.; Morton, R.E.; et al. Δ-5 Fatty Acid Desaturase FADS1 Impacts Metabolic Disease by Balancing Proinflammatory and Proresolving Lipid Mediators. Arterioscler. Thromb. Vasc Biol. 2018, 38, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Young, K.E.; Flaherty, S.; Woodman, K.M.; Sharma-Walia, N.; Reynolds, J.M. Fatty acid synthase regulates the pathogenicity of Th17 cells. J Leukoc. Biol. 2017, 102, 1229–1235. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vergara, D.; Nigro, A.; Romano, A.; De Domenico, S.; Damato, M.; Franck, J.; Coricciati, C.; Wistorski, M.; Cardon, T.; Fournier, I.; et al. Distinct Protein Expression Networks are Activated in Microglia Cells after Stimulation with IFN-γ and IL-4. Cells 2019, 8, 580. https://doi.org/10.3390/cells8060580
Vergara D, Nigro A, Romano A, De Domenico S, Damato M, Franck J, Coricciati C, Wistorski M, Cardon T, Fournier I, et al. Distinct Protein Expression Networks are Activated in Microglia Cells after Stimulation with IFN-γ and IL-4. Cells. 2019; 8(6):580. https://doi.org/10.3390/cells8060580
Chicago/Turabian StyleVergara, Daniele, Annamaria Nigro, Alessandro Romano, Stefania De Domenico, Marina Damato, Julien Franck, Chiara Coricciati, Maxence Wistorski, Tristan Cardon, Isabelle Fournier, and et al. 2019. "Distinct Protein Expression Networks are Activated in Microglia Cells after Stimulation with IFN-γ and IL-4" Cells 8, no. 6: 580. https://doi.org/10.3390/cells8060580
APA StyleVergara, D., Nigro, A., Romano, A., De Domenico, S., Damato, M., Franck, J., Coricciati, C., Wistorski, M., Cardon, T., Fournier, I., Quattrini, A., Salzet, M., Furlan, R., & Maffia, M. (2019). Distinct Protein Expression Networks are Activated in Microglia Cells after Stimulation with IFN-γ and IL-4. Cells, 8(6), 580. https://doi.org/10.3390/cells8060580