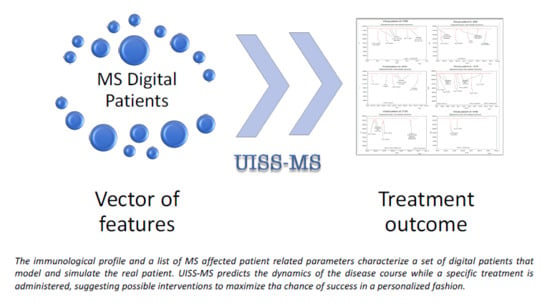
The Potential of Computational Modeling to Predict Disease Course and Treatment Response in Patients with Relapsing Multiple Sclerosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Computational Model
2.2. Introduction to the Simulation Framework
- Newborn cells are introduced in the resting state.
- A cell becomes active when it is stimulated through an interaction with another entity. For example, TH cell activation occurs with the interaction with an antigen if TCR binds the antigen.
- Anergic state. In this state, the cell does not interact. This state applies, for example, to B, TH, and TC cells.
- Status intern applies only to antigen-presenting cells (APC). When an APC (i.e., an M, DC, or B cells) encounters the antigen, it may be directed recognized by membrane-bound receptors, for example, those on the surface of a naive B cell. Unlike B cells, T cells do not recognize antigens directly. They “see” antigen as peptides only in association with the host surface MHC molecules. Since MHC molecules can only bind peptide molecules of 7–15 amino acids long, T cells only recognize their specific antigen in the form of short peptides. The antigen presenting cells such as dendritic cells and B-cells take up antigen and partially degrade it into peptides, which then occupy the antigen-presenting groove in MHC class I and MHC class II molecules.
- When cells present peptides via MHC molecules, their status change to PresI (MHC class I) or PresII (MHC class II).
- Status duplicating (applies to TH, TC, and B cells) is achieved when a cell has been activated and stimulated to start the clonal division.
- BoundToAb status applies to specific cells (for example, pathogen-infected cells). This state represents the fact that a cell has been recognized by an antibody (Ab). Cells in this status may die by the action of Ab-complement or through natural killer cells.
- Status infected applies to virus target cells. It identifies viral penetration via permeabilization of the host cell membrane when it begins local replication and spreads. Each cell can be in different internal states and all cells are tracked individually throughout an experimental run. This status can be actively or silently infected, according to the fact that the virus is replicating or not.
- Epithelial cell-pathogen. If an epithelial cell encounters, for example, a specific virus or intracellular bacteria, the cell is infected by the pathogen (status infected), then the cell may also present its infected status (PresI) to the immune system cells. Epithelial cells are reported here as an example, hence we can have different cells representing the different targets of different pathogens.
- IG_Ag. If a soluble immunoglobulin (IG) encounters its specific antigen (Ag), the IG binds to Ag and forms an immunocomplex (a macrophage can capture that). The binding probability is, as already said, a function of the Hamming distance of the two entities.
- B_Antigen. If a naive B lymphocyte expresses at the cell surface a membrane IG, which is specific for the native antigen (calculated according to the Hamming distance between the two strings), the B lymphocyte internalizes the membrane IG and the bound Ag (state intern). It then processes the IG–Ag complex into peptides, which are then presented by MHC-II (status PresII) at the B lymphocyte surface. We recall here that the binding probability is a function of the Hamming distance of the B receptor and the peptide (specific interaction). The B cell is now an APC (specialized antigen-presenting cell).
- M_Antigen. If a macrophage encounters a native antigen or an immunocomplex, the macrophage internalizes the antigen or the immunocomplex (state intern). Then it processes it into peptides, which are then presented by MHC-II (status PresII) at the macrophage cell surface. The M is now an APC (specialized antigen-presenting cell).
- DC_Antigen. If a naive dendritic cell encounters a native antigen or an immunocomplex, the dendritic cell internalizes the antigen or the immunocomplex (state intern), and then it processes it into peptides, which are then presented by MHC-II (status PresII) at the dendritic cell surface. The DC cell is now a specialized antigen-presenting cell (APC).
- TH_B. If a resting T helper lymphocyte (TH) encounters the B lymphocyte that is presenting a given peptide-MHC-II complex (status PresII), the TH cell becomes an activated T helper lymphocyte (status active) that helps the B cell to differentiate into the plasma cell or memory cell. At the molecular level, the interaction holds if the T cell receptor (TR) at the surface of the Th cells binds specifically to the peptide-MHC-II complex (specific interaction). Then active Th proliferates and secretes interleukin 2 (IL2). At the same time, B lymphocyte proliferates and differentiates into a plasma cell (that secretes IG) or into a memory cell (with IG at its surface).
- TH-M and TH-DC. As already shown in the TH_B interaction. If a resting TH encounters a macrophage or dendritic cell in the PresII state, TH becomes activated and secretes interleukins that activate other cells of the immune response (NK, mast cells, cytotoxic T lymphocytes, and others).
- TC–epithelial cell. If cytotoxic resting T lymphocyte (TC) encounters an epithelial cell that is infected (and thus also PresI status), for example, by a virus or intracellular bacteria, the cytotoxic T cell becomes, in the presence of IL2, an active cytotoxic T lymphocyte that kills the other cell. At the molecular level, the T cell receptor (TR) at the surface of a resting cytotoxic T lymphocyte binds specifically to the peptide-MHC-I complex at the surface of the cell. It must be noted that such interaction with TC cells may also arise for other infected cells such as, for example, M infected by tuberculosis.
- IG–epithelial cell (IG–bacteria, IG–virus). If an immunoglobulin IG encounters an infected epithelial cell that is presenting the antigen at its cell surface, and the soluble IG recognizes specifically the antigen, the opsonized cell (cell with bound IG on its surface) may be killed by complement-dependent cytotoxicity (CDC) or by antibody-dependent cell cytotoxicity (ADCC) mechanisms. At the molecular level, the first interaction is the recognition by the IG of the antigen expressed at the surface of the bacteria. A similar specific interaction with IG may also arise in other scenarios that include binding of IG to cancer cells, infected cells, viruses, bacteria, and others.
2.3. Extension of the Simulation Framework to Include MS Pathogenesis and Related Treatments
2.4. Simulation of Real Patients
3. Results
3.1. Prediction Robustness
3.2. Real Patients’ Predictions
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Yadav, S.K.; Mindur, J.E.; Ito, K.; Dhib-Jalbut, S. Advances in the immunopathogenesis of multiple sclerosis. Curr. Opin. Neurol. 2015, 28, 206–219. [Google Scholar] [CrossRef]
- Dendrou, C.A.; Fugger, L.; Friese, M.A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 2015, 15, 545–558. [Google Scholar] [CrossRef]
- Harding, K.; Williams, O.; Willis, M.; Hrastelj, J.; Rimmer, A.; Joseph, F.; Tomassini, V.; Wardle, M.; Pickersgill, T.; Robertson, N.; et al. Clinical Outcomes of Escalation vs. Early Intensive Disease-Modifying Therapy in Patients with Multiple Sclerosis. JAMA Neurol. 2019, 76, 536–541. [Google Scholar] [CrossRef]
- Trapp, B.D.; Peterson, J.; Ransohoff, R.M.; Rudick, R.; Mörk, S.; Bö, L. Axonal transection in the lesions of multiple sclerosis. N. Engl. J. Med. 1998, 338, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Filippi, M.; Rovaris, M.; Inglese, M.; Barkhof, F.; De Stefano, N.; Smith, S.; Comi, P.G. Interferon beta-1a for brain tissue loss in patients at presentation with syndromes suggestive of multiple sclerosis: A randomised, double-blind, placebo-controlled trial. Lancet 2004, 364, 1489–1496. [Google Scholar] [CrossRef]
- Rojas, J.; Patrucco, L.; Cristiano, E. Brain atrophy in radiologically isolated syndromes. J. Neuroimaging 2014, 25. [Google Scholar] [CrossRef] [PubMed]
- Knier, B.; Berthele, A.; Buck, D.; Schmidt, P.; Zimmer, C.; Mühlau, M.; Hemmer, B.; Korn, T. Optical coherence tomography indicates disease activity prior to clinical onset of central nervous system demyelination. Mult. Scler. 2016, 22, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Ziemssen, T.; De Stefano, N.; Sormani, M.P.; Van Wijmeersch, B.; Wiendl, H.; Kieseier, B.C. Optimizing therapy early in multiple sclerosis: An evidence-based view. Mult. Scler. Relat. Disord. 2015, 4, 460–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havrdova, E.; Galetta, S.; Hutchinson, M.; Stefoski, D.; Bates, D.; Polman, C.H.; O’Connor, P.W.; Giovannoni, G.; Phillips, J.T.; Lublin, F.D.; et al. Effect of natalizumab on clinical and radiological disease activity in multiple sclerosis: A retrospective analysis of the Natalizumab Safety and Efficacy in Relapsing-Remitting Multiple Sclerosis (AFFIRM) study. Lancet Neurol. 2009, 8, 254–260. [Google Scholar] [CrossRef]
- Rae-Grant, A.; Day, G.S.; Marrie, R.A.; Rabinstein, A.; Cree, B.A.C.; Gronseth, G.S.; Haboubi, M.; Halper, J.; Hosey, J.P.; Jones, D.E.; et al. Practice guideline recommendations summary: Disease-modifying therapies for adults with multiple sclerosis. Neurology 2018, 90, 777–788. [Google Scholar] [CrossRef] [Green Version]
- Lünemann, J.D.; Ruck, T.; Muraro, P.A.; Bar’Or, A.; Wiendl, H. Immune reconstitution therapies: Concepts for durable remission in multiple sclerosis. Nat. Rev. Neurol. 2019. Available online: https://www.nature.com/articles/s41582-019-0268-z (accessed on 23 February 2020).
- Pennisi, M.; Rajput, A.-M.; Toldo, L.; Pappalardo, F. Agent based modeling of Treg-Teff cross regulation in relapsing-remitting multiple sclerosis. BMC Bioinform. 2013, 14, S9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pappalardo, F.; Rajput, A.-M.; Motta, S. Computational modeling of brain pathologies: The case of multiple sclerosis. Brief. Bioinform. 2018, 19, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Vélez de Mendizábal, N.; Carneiro, J.; Solé, R.V.; Goñi, J.; Bragard, J.; Martinez-Forero, I.; Martinez-Pasamar, S.; Sepulcre, J.; Torrealdea, J.; Bagnato, F.; et al. Modeling the effector - regulatory T cell cross-regulation reveals the intrinsic character of relapses in Multiple Sclerosis. BMC Syst. Biol. 2011, 5, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pappalardo, F.; Pennisi, M.; Rajput, A.M.; Chiacchio, F.; Motta, S. Relapsing-remitting multiple scleroris and the role of vitamin D: An agent based model. In Proceedings of the ACM BCB 2014—5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics. 2014. Available online: https://doi.org/10.1145/2649387.2660844 (accessed on 23 February 2020).
- Pennisi, M.; Russo, G.; Motta, S.; Pappalardo, F. Agent based modeling of the effects of potential treatments over the blood–brain barrier in multiple sclerosis. J. Immunol. Methods 2015, 427, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Beccuti, M.; Cazzaniga, P.; Pennisi, M.; Besozzi, D.; Nobile, M.S.; Pernice, S.; Russo, G.; Tangherloni, A.; Pappalardo, F. GPU Accelerated Analysis of Treg-Teff Cross Regulation in Relapsing-Remitting Multiple Sclerosis. In Euro-Par 2018 Workshops, LNCS 11339; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2019; Volume 11339, pp. 626–637. ISBN 978-3-642-54419-4. [Google Scholar]
- Pernice, S.; Beccuti, M.; Do, P.; Pennisi, M.; Pappalardo, F. Estimating Daclizumab effects in Multiple Sclerosis using Stochastic Symmetric Nets. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine, BIBM: Location of Conference, Madrid, Spain, 3–6 December 2018. [Google Scholar]
- Pernice, S.; Pennisi, M.; Romano, G.; Maglione, A.; Cutrupi, S.; Pappalardo, F.; Balbo, G.; Beccuti, M.; Cordero, F.; Calogero, R.A. A computational approach based on the Colored Petri Net formalism for studying Multiple Sclerosis. BMC Bioinform. 2019, 20, 1–17. [Google Scholar] [CrossRef]
- Pernice, S.; Romano, G.; Russo, G.; Beccuti, M.; Pappalardo, F. Exploiting Stochastic Petri Net formalism to capture the Relapsing Remitting Multiple Sclerosis variability under Daclizumab administration. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019. [Google Scholar]
- Kannan, V.; Kiani, N.A.; Piehl, F.; Tegner, J. A minimal unified model of disease trajectories captures hallmarks of multiple sclerosis. Math. Biosci. 2017, 289, 1–8. [Google Scholar] [CrossRef]
- Kotelnikova, E.; Kiani, N.A.; Abad, E.; Martinez-Lapiscina, E.H.; Andorra, M.; Zubizarreta, I.; Pulido-Valdeolivas, I.; Pertsovskaya, I.; Alexopoulos, L.G.; Olsson, T.; et al. Dynamics and heterogeneity of brain damage in multiple sclerosis. PLoS Comput. Biol. 2017, 13, e1005757. [Google Scholar] [CrossRef] [Green Version]
- Malhotra, A.; Gündel, M.; Rajput, A.M.; Mevissen, H.-T.; Saiz, A.; Pastor, X.; Lozano-Rubi, R.; Martinez-Lapsicina, E.H.; Zubizarreta, I.; Mueller, B.; et al. Knowledge Retrieval from PubMed Abstracts and Electronic Medical Records with the Multiple Sclerosis Ontology. PLoS ONE 2015, 10, e0116718. [Google Scholar] [CrossRef]
- Musen, M.A. The protégé project. AI Matters 2015, 1, 4–12. [Google Scholar] [CrossRef]
- Read, M.; Andrews, P.S.; Timmis, J.; Kumar, V. Modelling biological behaviours with the unified modelling language: An immunological case study and critique. J. R. Soc. Interface 2014, 11, 20140704. [Google Scholar] [CrossRef]
- Palladini, A.; Nicoletti, G.; Pappalardo, F.; Murgo, A.; Grosso, V.; Stivani, V.; Ianzano, M.L.; Antognoli, A.; Croci, S.; Landuzzi, L.; et al. In silico modeling and in vivo efficacy of cancer-preventive vaccinations. Cancer Res. 2010, 70, 7755–7763. [Google Scholar] [CrossRef] [Green Version]
- Pappalardo, F.; Pennisi, M.; Ricupito, A.; Topputo, F.; Bellone, M. Induction of T-cell memory by a dendritic cell vaccine: A computational model. Bioinformatics 2014, 30, 1884–1891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pappalardo, F.; Fichera, E.; Paparone, N.; Lombardo, A.; Pennisi, M.; Russo, G.; Leotta, M.; Pappalardo, F.; Pedretti, A.; De Fiore, F.; et al. A computational model to predict the immune system activation by citrus-derived vaccine adjuvants. Bioinformatics 2016, 32, 2672–2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennisi, M.; Pappalardo, F.; Motta, S. Agent Based Modeling of Lung Metastasis-Immune System Competition. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Andrews, P.S., Timmis, J., Owens, N.D.L., Aickelin, U., Hart, E., Hone, A., Tyrrell, A.M., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2009; Volume 5666, pp. 1–3. ISBN 978-3-642-03245-5. [Google Scholar]
- Pappalardo, F.; Motta, S.; Lollini, P.-L.; Mastriani, E. Analysis of vaccine’s schedules using models. Cell. Immunol. 2006, 244, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Celada, F.; Seiden, P.E. A computer model of cellular interactions in the immune system. Immunol. Today 1992, 13, 56–62. [Google Scholar] [CrossRef]
- Farmer, J.D.; Packard, N.H.; Perelson, A.S. The immune system, adaptation, and machine learning. Phys. D Nonlinear Phenom. 1986, 22, 187–204. [Google Scholar] [CrossRef]
- Pennisi, M.; Russo, G.; Ravalli, S.; Pappalardo, F. Combining agent based-models and virtual screening techniques to predict the best citrus-derived vaccine adjuvants against human papilloma virus. BMC Bioinform. 2017, 18. [Google Scholar] [CrossRef]
- Sospedra, M.; Martin, R. Immunology of Multiple Sclerosis. Semin. Neurol. 2016, 36, 115–127. [Google Scholar] [CrossRef]
- Pappalardo, F.; Russo, G.; Pennisi, M.; Sgroi, G.; Parasiliti Palumbo, G.A.; Motta, S.; Maimone, D.; Chiacchio, F. Agent based modeling of relapsing multiple sclerosis: A possible approach to predict treatment outcome. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 1380–1385. [Google Scholar]
- Jeker, L.T.; Bour-Jordan, H.; Bluestone, J.A. Breakdown in Peripheral Tolerance in Type 1 Diabetes in Mice and Humans. Cold Spring Harb. Perspect. Med. 2012, 2, a007807. [Google Scholar] [CrossRef] [Green Version]
- Oskari Virtanen, J.; Jacobson, S. Viruses and Multiple Sclerosis. CNS Neurol. Disord-Drug Targets 2012, 11, 528–544. [Google Scholar] [CrossRef] [Green Version]
- Monzani, F. Review of the clinical evidence for interferon interferon beta 1a (Rebif) in the treatment of multiple sclerosis. Neuropsychiatr. Dis. Treat. 2008, 4, 321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas Abul, K.; Lichtman, A.H.; Pillai, S. Cellular and Molecular Immunology, 8th ed.; Elsevier: Amsterdam, The Netherlands, 2014; ISBN 8535259724. [Google Scholar]
- Dhib-Jalbut, S. Mechanisms of action of interferons and glatiramer acetate in multiple sclerosis. Neurology 2002, 58, S3–S9. [Google Scholar] [CrossRef] [PubMed]
- de Andrés, C.; Aristimuño, C.; de las Heras, V.; Martínez-Ginés, M.L.; Bartolomé, M.; Arroyo, R.; Navarro, J.; Giménez-Roldán, S.; Fernández-Cruz, E.; Sánchez-Ramón, S. Interferon beta-1a therapy enhances CD4+ regulatory T-cell function: An ex vivo and in vitro longitudinal study in relapsing−remitting multiple sclerosis. J. Neuroimmunol. 2007, 182, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Bar-Or, A.; Pachner, A.; Menguy-Vacheron, F.; Kaplan, J.; Wiendl, H. Teriflunomide and Its Mechanism of Action in Multiple Sclerosis. Drugs 2014, 74, 659–674. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, 277–280. [Google Scholar] [CrossRef] [Green Version]
- Hoops, S.; Sahle, S.; Gauges, R.; Lee, C.; Pahle, J.; Simus, N.; Singhal, M.; Xu, L.; Mendes, P.; Kummer, U. COPASI--a COmplex PAthway SImulator. Bioinformatics 2006, 22, 3067–3074. [Google Scholar] [CrossRef] [Green Version]
- Ingwersen, J.; Aktas, O.; Kuery, P.; Kieseier, B.; Boyko, A.; Hartung, H.-P. Fingolimod in multiple sclerosis: Mechanisms of action and clinical efficacy. Clin. Immunol. 2012, 142, 15–24. [Google Scholar] [CrossRef]
- Rice, G.P.A.; Hartung, H.-P.; Calabresi, P.A. Anti- 4 integrin therapy for multiple sclerosis: Mechanisms and rationale. Neurology 2005, 64, 1336–1342. [Google Scholar] [CrossRef]
- Miller, D.H.; Soon, D.; Fernando, K.T.; MacManus, D.G.; Barker, G.J.; Yousry, T.A.; Fisher, E.; O’Connor, P.W.; Phillips, J.T.; Polman, C.H.; et al. MRI outcomes in a placebo-controlled trial of natalizumab in relapsing MS. Neurology 2007, 68, 1390–1401. [Google Scholar] [CrossRef]
- Polman, C.H.; O’Connor, P.W.; Havrdova, E.; Hutchinson, M.; Kappos, L.; Miller, D.H.; Phillips, J.T.; Lublin, F.D.; Giovannoni, G.; Wajgt, A.; et al. A Randomized, Placebo-Controlled Trial of Natalizumab for Relapsing Multiple Sclerosis. N. Engl. J. Med. 2006, 354, 899–910. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, P.S.; Blinkenberg, M. The potential role for ocrelizumab in the treatment of multiple sclerosis: Current evidence and future prospects. Ther. Adv. Neurol. Disord. 2016, 9, 44–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalincik, T.; Manouchehrinia, A.; Sobisek, L.; Jokubaitis, V.; Spelman, T.; Horakova, D.; Havrdova, E.; Trojano, M.; Izquierdo, G.; Lugaresi, A.; et al. Towards personalized therapy for multiple sclerosis: Prediction of individual treatment response. Brain 2017, 140, 2426–2443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tintore, M.; Rovira, À.; Río, J.; Otero-Romero, S.; Arrambide, G.; Tur, C.; Comabella, M.; Nos, C.; Arévalo, M.J.; Negrotto, L.; et al. Defining high, medium and low impact prognostic factors for developing multiple sclerosis. Brain 2015, 138, 1863–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correale, J.; De los Milagros Bassani Molinas, M. Oligoclonal bands and antibody responses in Multiple Sclerosis. J. Neurol. 2002, 249, 375–389. [Google Scholar] [CrossRef]
- Bernitsas, E.; Khan, O.; Razmjou, S.; Tselis, A.; Bao, F.; Caon, C.; Millis, S.; Seraji-Bozorgzad, N. Cerebrospinal fluid humoral immunity in the differential diagnosis of multiple sclerosis. PLoS ONE 2017, 12, e0181431. [Google Scholar] [CrossRef] [Green Version]
- Gold, S.M.; Voskuhl, R.R. Pregnancy and multiple sclerosis: From molecular mechanisms to clinical application. Semin. Immunopathol. 2016, 38, 709–718. [Google Scholar] [CrossRef]
- Voskuhl, R.; Momtazee, C. Pregnancy: Effect on Multiple Sclerosis, Treatment Considerations, and Breastfeeding. Neurotherapeutics 2017, 14, 974–984. [Google Scholar] [CrossRef]
- Rotstein, D.; Montalban, X. Reaching an evidence-based prognosis for personalized treatment of multiple sclerosis. Nat. Rev. Neurol. 2019, 15, 287–300. [Google Scholar] [CrossRef]
- Olsen, J.A.; Kenna, L.A.; Tipon, R.C.; Spelios, M.G.; Stecker, M.M.; Akirav, E.M. A Minimally-invasive Blood-derived Biomarker of Oligodendrocyte Cell-loss in Multiple Sclerosis. EBioMedicine 2016, 10, 227–235. [Google Scholar] [CrossRef] [Green Version]





| Entity | Initial Quantity per µL (or Cubic Millimeter) | Half-Life (Days or Hours) |
|---|---|---|
| B | 260 | 3.3 days |
| TH | 876 | 3.3 days |
| TC | 434 | 3.3 days |
| CDC | 351 | 3.3 days |
| M | 351 | 3.3 days |
| P | 0 | 3.3 days |
| IC | 0 | 4.0 days |
| ODC | 45,000 | 4.0 days |
| IFN-G | 0 | 1.6 days |
| IL-2 | 0 | 1.6 days |
| IL-4 | 0 | 1.6 days |
| IL-10 | 0 | 1.6 days |
| IL-12 | 0 | 1.6 days |
| IL-17 | 0 | 1.6 days |
| IL-23 | 0 | 1.6 days |
| TGFB | 0 | 1.6 days |
| MBP (myelin basic protein) | 0 | 3 days |
| IgG | 0 | 23.0 days |
| Chemokine (generic) | 0 | 3.0 h |
| Patient ID | Age Range | MS Age Onset | Oligoclonal Bands (OB) | Pregnancy | Lesional Load (YYYY/MM–Number) | Spinal Lesions (YYYY/MM–Number) | Treatment |
|---|---|---|---|---|---|---|---|
| 1070 | 20–29 | 20 | yes | no | 2007/03–32007/06–5 2010/10–77 2010/12–82 2013/05–59 2015/06–41 | 2013/05–6 2015/06–7 | 2007/09: IFN-β1a 2013/05: fingolimod 2015/12: natalizumab |
| 3736 | 30–39 | 30 | yes | yes, from 2015/11 to 2016/08 | 2014/09–22 2015/01–28 | N/D | 2015/02-2015/12: natalizumab 2016/09: natalizumab |
| 2789 | 30–39 | 33 | yes | yes, from 2012/10 to 2013/07 and from 2015/11 to 2016/08 | 2013/11–9 2013/12–9 2014/09–9 2017/01–14 | N/D | 2017/02: teriflunomide |
| 5793 | 20–29 | 26 | yes | no | 2015/08–20 2015/09–22 2016/01–28 | N/D | 2016/06: natalizumab |
| 2070 | 30–39 | 31 | no | no | 2014/10–23 2015/03–37 2016/01–33 | N/D | 2014/12: IFN-β1a |
| 2961 | 30–39 | 29 | no | no | 2015/03–49 2016/03–43 2017/04–65 | N/D | N/A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pappalardo, F.; Russo, G.; Pennisi, M.; Parasiliti Palumbo, G.A.; Sgroi, G.; Motta, S.; Maimone, D. The Potential of Computational Modeling to Predict Disease Course and Treatment Response in Patients with Relapsing Multiple Sclerosis. Cells 2020, 9, 586. https://doi.org/10.3390/cells9030586
Pappalardo F, Russo G, Pennisi M, Parasiliti Palumbo GA, Sgroi G, Motta S, Maimone D. The Potential of Computational Modeling to Predict Disease Course and Treatment Response in Patients with Relapsing Multiple Sclerosis. Cells. 2020; 9(3):586. https://doi.org/10.3390/cells9030586
Chicago/Turabian StylePappalardo, Francesco, Giulia Russo, Marzio Pennisi, Giuseppe Alessandro Parasiliti Palumbo, Giuseppe Sgroi, Santo Motta, and Davide Maimone. 2020. "The Potential of Computational Modeling to Predict Disease Course and Treatment Response in Patients with Relapsing Multiple Sclerosis" Cells 9, no. 3: 586. https://doi.org/10.3390/cells9030586
APA StylePappalardo, F., Russo, G., Pennisi, M., Parasiliti Palumbo, G. A., Sgroi, G., Motta, S., & Maimone, D. (2020). The Potential of Computational Modeling to Predict Disease Course and Treatment Response in Patients with Relapsing Multiple Sclerosis. Cells, 9(3), 586. https://doi.org/10.3390/cells9030586