Microautophagy in Plants: Consideration of Its Molecular Mechanism
Abstract
:1. Introduction
2. Microautophagy in Yeast and Animals
2.1. Microautophagy in Yeast—a Model for Plant Studies
2.2. Regulatory Factors
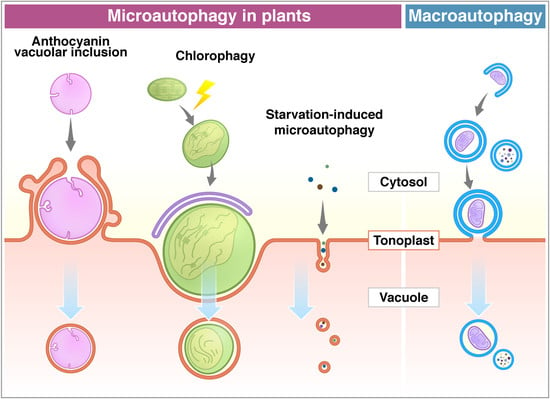
3. Microautophagy in Plants
3.1. Microautophagy Reported in Plants
3.2. Methods for Microautophagy Observation
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011, 66, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Oikawa, K.; Yoshimoto, K.; Kondo, M.; Mano, S.; Yamada, K.; Hayashi, M.; Sakamoto, W.; Ohsumi, Y.; Nishimura, M. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 2013, 25, 4967–4983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Mugume, Y.; Bassham, D.C. New advances in autophagy in plants: Regulation, selectivity and function. Semin. Cell Dev. Biol. 2018, 80, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.; Tarkowski, L.P.; Van den Ende, W.; Bassham, D.C. Linking Autophagy to Abiotic and Biotic Stress Responses. Trends Plant Sci. 2019, 24, 413–430. [Google Scholar] [CrossRef] [Green Version]
- Van Doorn, W.G.; Papini, A. Ultrastructure of autophagy in plant cells: A review. Autophagy 2013, 9, 1922–1936. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, C.; Legouis, R.; Culetto, E. ESCRT and autophagies: Endosomal functions and beyond. Semin. Cell Dev. Biol. 2018, 74, 21–28. [Google Scholar] [CrossRef]
- Ohsumi, Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [Green Version]
- De Duve, C.; Wattiaux, R. Functions of lysosomes. Annu. Rev. Physiol. 1966, 28, 435–492. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Schuck, S.; Gallagher, C.M.; Walter, P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 2014, 127, 4078–4088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oku, M.; Sakai, Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries. Bioessays 2018, 40, e1800008. [Google Scholar] [CrossRef] [PubMed]
- Müller, O.; Sattler, T.; Flötenmeyer, M.; Schwarz, H.; Plattner, H.; Mayer, A. Autophagic tubes: Vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J. Cell Biol. 2000, 151, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.; Kaushik, S.; Clement, C.C.; Cannizzo, E.S.; Scharf, B.; Follenzi, A.; Potolicchio, I.; Nieves, E.; Cuervo, A.M.; Santambrogio, L. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 2011, 20, 131–139. [Google Scholar] [CrossRef] [Green Version]
- Tamura, N.; Oku, M.; Sakai, Y. Atg21 regulates pexophagy via its PI(3)P-binding activity in Pichia pastoris. FEMS Yeast Res. 2014, 14, 435–444. [Google Scholar] [CrossRef] [Green Version]
- Mukaiyama, H.; Baba, M.; Osumi, M.; Aoyagi, S.; Kato, N.; Ohsumi, Y.; Sakai, Y. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell 2004, 15, 58–70. [Google Scholar] [CrossRef]
- Ano, Y.; Hattori, T.; Oku, M.; Mukaiyama, H.; Baba, M.; Ohsumi, Y.; Kato, N.; Sakai, Y. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol-3-phosphate. Mol. Biol. Cell 2005, 16, 446–457. [Google Scholar] [CrossRef]
- Roberts, P.; Moshitch-Moshkovitz, S.; Kvam, E.; O’Toole, E.; Winey, M.; Goldfarb, D.S. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 129–141. [Google Scholar] [CrossRef] [Green Version]
- Krick, R.; Muehe, Y.; Prick, T.; Bremer, S.; Schlotterhose, P.; Eskelinen, E.L.; Millen, J.; Goldfarb, D.S.; Thumm, M. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell 2008, 19, 4492–4505. [Google Scholar] [CrossRef] [Green Version]
- Lemasters, J.J. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3). Redox Biol. 2014, 2, 749–754. [Google Scholar] [CrossRef] [Green Version]
- Vevea, J.D.; Garcia, E.J.; Chan, R.B.; Zhou, B.; Schultz, M.; Di Paolo, G.; McCaffery, J.M.; Pon, L.A. Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 2015, 35, 584–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Zutphen, T.; Todde, V.; de Boer, R.; Kreim, M.; Hofbauer, H.F.; Wolinski, H.; Veenhuis, M.; van der Klei, I.J.; Kohlwein, S.D. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2014, 25, 290–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, T.; Fujimoto, M.; Tatematsu, T.; Cheng, J.; Orii, M.; Takatori, S.; Fujimoto, T. Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, D.L.; Dunn, W.A. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 1995, 108, 25–35. [Google Scholar] [PubMed]
- Dunn, W.A., Jr.; Cregg, J.M.; Kiel, J.A.; van der Klei, I.J.; Oku, M.; Sakai, Y.; Sibirny, A.A.; Stasyk, O.V.; Veenhuis, M. Pexophagy: The selective autophagy of peroxisomes. Autophagy 2005, 1, 75–83. [Google Scholar] [CrossRef] [Green Version]
- Sakai, Y.; Koller, A.; Rangell, L.K.; Keller, G.A.; Subramani, S. Peroxisome degradation by microautophagy in Pichia pastoris: Identification of specific steps and morphological intermediates. J. Cell Biol. 1998, 141, 625–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Goldfarb, D.S. YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. J. Cell Sci. 1998, 111, 2137–2147. [Google Scholar]
- Schuck, S.; Prinz, W.A.; Thorn, K.S.; Voss, C.; Walter, P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 2009, 187, 525–536. [Google Scholar] [CrossRef] [Green Version]
- Schafer, J.A.; Schessner, J.P.; Bircham, P.W.; Tsuji, T.; Funaya, C.; Pajonk, O.; Schaeff, K.; Ruffini, G.; Papagiannidis, D.; Knop, M.; et al. ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. EMBO J. 2020, 39, e102586. [Google Scholar] [CrossRef]
- Oku, M.; Maeda, Y.; Kagohashi, Y.; Kondo, T.; Yamada, M.; Fujimoto, T.; Sakai, Y. Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 2017, 216, 3263–3274. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.W.; Li, J.; Bao, J.K. Microautophagy: Lesser-known self-eating. Cell Mol. Life Sci. 2012, 69, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Patel, B.; Koga, H.; Cuervo, A.M.; Jenny, A. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy 2016, 12, 1984–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamagata, M.; Obara, K.; Kihara, A. Sphingolipid synthesis is involved in autophagy in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2011, 410, 786–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stromhaug, P.E.; Bevan, A.; Dunn, W.A., Jr. GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in Pichia pastoris. J. Biol. Chem. 2001, 276, 42422–42435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukaiyama, H.; Oku, M.; Baba, M.; Samizo, T.; Hammond, A.T.; Glick, B.S.; Kato, N.; Sakai, Y. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells 2002, 7, 75–90. [Google Scholar] [CrossRef]
- Nazarko, V.Y.; Nazarko, T.Y.; Farre, J.C.; Stasyk, O.V.; Warnecke, D.; Ulaszewski, S.; Cregg, J.M.; Sibirny, A.A.; Subramani, S. Atg35, a micropexophagy-specific protein that regulates micropexophagic apparatus formation in Pichia pastoris. Autophagy 2011, 7, 375–385. [Google Scholar] [CrossRef] [Green Version]
- Yoshimoto, K. Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol. 2012, 53, 1355–1365. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Chung, T.; Vierstra, R.D. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 2014, 26, 788–807. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.N.; Zarza, X.; Kim, J.H.; Yoon, M.J.; Kim, S.H.; Lee, J.H.; Paris, N.; Munnik, T.; Otegui, M.S.; Chung, T. Vacuolar trafficking protein VPS38 is dispensable for autophagy. Plant Physiol. 2018, 176, 1559–1572. [Google Scholar] [CrossRef]
- Sattler, T.; Mayer, A. Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J. Cell Biol. 2000, 151, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Oku, M.; Sakai, Y. Pexophagy in yeasts. Biochim. Biophys. Acta 2016, 1863, 992–998. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, E.S.; Choi, Y.; Hwang, I.; Staiger, C.J.; Chung, Y.Y.; Lee, Y. The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol. 2008, 147, 1886–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, N.; Gao, X.Q.; Zhao, X.Y.; Zhu, D.Z.; Zhou, L.Z.; Zhang, X.S. Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation. Plant Mol. Biol. 2011, 77, 251–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldman, M.J.; Poirier, B.C.; Lange, B.M. Misexpression of the Niemann-Pick disease type C1 (NPC1)-like protein in Arabidopsis causes sphingolipid accumulation and reproductive defects. Planta 2015, 242, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Uttenweiler, A.; Schwarz, H.; Neumann, H.; Mayer, A. The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol. Biol. Cell 2007, 18, 166–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toulmay, A.; Prinz, W.A. Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 2013, 202, 35–44. [Google Scholar] [CrossRef]
- Gao, C.; Zhuang, X.; Shen, J.; Jiang, L. Plant ESCRT complexes: Moving beyond endosomal sorting. Trends Plant Sci. 2017, 22, 986–998. [Google Scholar] [CrossRef]
- Korbei, B.; Moulinier-Anzola, J.; De-Araujo, L.; Lucyshyn, D.; Retzer, K.; Khan, M.A.; Luschnig, C. Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr. Biol. 2013, 23, 2500–2505. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Luo, M.; Zhao, Q.; Yang, R.; Cui, Y.; Zeng, Y.; Xia, J.; Jiang, L. A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr. Biol. 2014, 24, 2556–2563. [Google Scholar] [CrossRef] [Green Version]
- Reyes, F.C.; Buono, R.A.; Roschzttardtz, H.; Di Rubbo, S.; Yeun, L.H.; Russinova, E.; Otegui, M.S. A novel endosomal sorting complex required for transport (ESCRT) component in Arabidopsis thaliana controls cell expansion and development. J. Biol. Chem. 2014, 289, 4980–4988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eastmond, P.J.; Quettier, A.L.; Kroon, J.T.; Craddock, C.; Adams, N.; Slabas, A.R. Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 2010, 22, 2796–2811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitakura, S.; Vanneste, S.; Robert, S.; Lofke, C.; Teichmann, T.; Tanaka, H.; Friml, J. Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 2011, 23, 1920–1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebine, K.; Okatani, Y.; Uemura, T.; Goh, T.; Shoda, K.; Niihama, M.; Morita, M.T.; Spitzer, C.; Otegui, M.S.; Nakano, A.; et al. A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 2008, 20, 3006–3021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Han, S.W.; Rodriguez-Welsh, M.F.; Rojas-Pierce, M. Homotypic vacuole fusion requires VTI11 and is regulated by phosphoinositides. Mol. Plant 2014, 7, 1026–1040. [Google Scholar] [CrossRef] [Green Version]
- Takemoto, K.; Ebine, K.; Askani, J.C.; Kruger, F.; Gonzalez, Z.A.; Ito, E.; Goh, T.; Schumacher, K.; Nakano, A.; Ueda, T. Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc. Natl. Acad Sci. USA 2018, 115, E2457–E2466. [Google Scholar] [CrossRef] [Green Version]
- Brillada, C.; Zheng, J.; Kruger, F.; Rovira-Diaz, E.; Askani, J.C.; Schumacher, K.; Rojas-Pierce, M. Phosphoinositides control the localization of HOPS subunit VPS41, which together with VPS33 mediates vacuole fusion in plants. Proc. Natl. Acad Sci. USA 2018, 115, E8305–E8314. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.M.; Sun, L.L.; Hu, W.; Ding, Y.H.; Dong, M.Q.; Du, L.L. ESCRTs cooperate with a selective autophagy receptor to mediate vacuolar targeting of soluble cargos. Mol. Cell 2015, 59, 1035–1042. [Google Scholar] [CrossRef] [Green Version]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef]
- Uytterhoeven, V.; Lauwers, E.; Maes, I.; Miskiewicz, K.; Melo, M.N.; Swerts, J.; Kuenen, S.; Wittocx, R.; Corthout, N.; Marrink, S.J.; et al. Hsc70-4 deforms membranes to promote synaptic protein turnover by endosomal microautophagy. Neuron 2015, 88, 735–748. [Google Scholar] [CrossRef] [Green Version]
- Noel, L.D.; Cagna, G.; Stuttmann, J.; Wirthmuller, L.; Betsuyaku, S.; Witte, C.P.; Bhat, R.; Pochon, N.; Colby, T.; Parker, J.E. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 2007, 19, 4061–4076. [Google Scholar] [CrossRef] [Green Version]
- Lin, B.L.; Wang, J.S.; Liu, H.C.; Chen, R.W.; Meyer, Y.; Barakat, A.; Delseny, M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 201–208. [Google Scholar] [CrossRef]
- Pattingre, S.; Espert, L.; Biard-Piechaczyk, M.; Codogno, P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie 2008, 90, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bassham, D.C. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE 2010, 5, e11883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubouloz, F.; Deloche, O.; Wanke, V.; Cameroni, E.; De Virgilio, C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 2005, 19, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Yu, B.; Wang, Y.; Liu, Y. Role of plant autophagy in stress response. Protein Cell 2011, 2, 784–791. [Google Scholar] [CrossRef] [Green Version]
- Chanoca, A.; Kovinich, N.; Burkel, B.; Stecha, S.; Bohorquez-Restrepo, A.; Ueda, T.; Eliceiri, K.W.; Grotewold, E.; Otegui, M.S. Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant Cell 2015, 27, 2545–2559. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, S.; Hidema, J.; Sakamoto, W.; Ishida, H.; Izumi, M. Selective elimination of membrane-damaged chloroplasts via microautophagy. Plant Physiol. 2018, 177, 1007–1026. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, S.; Izumi, M. Chlorophagy is ATG gene-dependent microautophagy process. Plant Signal. Behav. 2019, 14, 1554469. [Google Scholar] [CrossRef] [Green Version]
- Goto-Yamada, S.; Oikawa, K.; Bizan, J.; Shigenobu, S.; Yamaguchi, K.; Mano, S.; Hayashi, M.; Ueda, H.; Hara-Nishimura, I.; Nishimura, M.; et al. Sucrose starvation induces microautophagy in plant root cells. Front. Plant Sci. 2019, 10, 1604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriyasu, Y.; Ohsumi, Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996, 111, 1233–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, Y.; Suzuki, T.; Hattori, M.; Yoshimoto, K.; Ohsumi, Y.; Moriyasu, Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 2006, 47, 1641–1652. [Google Scholar] [CrossRef] [PubMed]
- Hiratsuka, R.; Terasaka, O. Pollen tube reuses intracellular components of nucellar cells undergoing programmed cell death in Pinus densiflora. Protoplasma 2011, 248, 339–351. [Google Scholar] [CrossRef]
- Van Doorn, W.G.; Kirasak, K.; Ketsa, S. Amorphous areas in the cytoplasm of Dendrobium tepal cells: Production through organelle degradation and destruction through macroautophagy? Autophagy 2013, 9, 1159–1166. [Google Scholar] [CrossRef] [Green Version]
- Sobieszczuk-Nowicka, E.; Wrzesinski, T.; Bagniewska-Zadworna, A.; Kubala, S.; Rucinska-Sobkowiak, R.; Polcyn, W.; Misztal, L.; Mattoo, A.K. Physio-genetic dissection of dark-induced leaf senescence and timing its reversal in barley. Plant Physiol. 2018, 178, 654–671. [Google Scholar] [CrossRef] [Green Version]
- Betz, W.J.; Mao, F.; Smith, C.B. Imaging exocytosis and endocytosis. Curr. Opin. Neurobiol. 1996, 6, 365–371. [Google Scholar] [CrossRef]
- Bolte, S.; Talbot, C.; Boutte, Y.; Catrice, O.; Read, N.D.; Satiat-Jeunemaitre, B. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 2004, 214, 159–173. [Google Scholar] [CrossRef]
- Yamada, K.; Fuji, K.; Shimada, T.; Nishimura, M.; Hara-Nishimura, I. Endosomal proteases facilitate the fusion of endosomes with vacuoles at the final step of the endocytotic pathway. Plant J. 2005, 41, 888–898. [Google Scholar] [CrossRef]
- Scheuring, D.; Scholler, M.; Kleine-Vehn, J.; Lofke, C. Vacuolar staining methods in plant cells. Methods Mol. Biol. 2015, 1242, 83–92. [Google Scholar] [CrossRef]
- Takatsuka, C.; Inoue, Y.; Higuchi, T.; Hillmer, S.; Robinson, D.G.; Moriyasu, Y. Autophagy in tobacco BY-2 cells cultured under sucrose starvation conditions: Isolation of the autolysosome and its characterization. Plant Cell Physiol. 2011, 52, 2074–2087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merkulova, E.A.; Guiboileau, A.; Naya, L.; Masclaux-Daubresse, C.; Yoshimoto, K. Assessment and optimization of autophagy monitoring methods in Arabidopsis roots indicate direct fusion of autophagosomes with vacuoles. Plant Cell Physiol. 2014, 55, 715–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Factors Reported to be Involved in Microautophagy * | Roles in Microautophagy | Reference | Homologs in Arabidopsis | |
|---|---|---|---|---|
| ATG | Micropexophagy (yeast): Atg1–5, Atg7–9, Atg11, Atg16, Atg17, Atg18, Atg21, Atg23, Atg24, Atg26, Atg28, Atg30, Atg35 | Recognition of peroxisomes. Formation of the MIPA. Isolation of peroxisomes and transportation of the cargo into the vacuole. | [15,16,17,35,36,37] | ATG1–16, ATG18, ATG101 [38,39,40] |
| PMN (yeast): Atg1–18, Atg21, Atg24, Atg29, Atg31 | Enclosure of the terminal vacuole and fusion. | [18,19] | ||
| Microlipophagy (yeast): Atg1–10, Atg12, Atg14–18 | Involved in the internalization of lipid droplets. Degradation of lipids by vacuolar lipase Atg15. Involved in microdomain formation during microautophagy in the stationary phase and nitrogen starvation. Involved in proper NPC distribution on the vacuolar membrane. | [22,23,41] | ||
| PI3K complex ** | Vps15, Vps34 | Generate PI3P on the membrane to trigger microautophagy in micropexophagy, microlipophagy. | [16,22,42] | VPS15, VPS34 [43,44] |
| Vacuolar membrane protein | Vac8 | Involved in vacuolar membrane fusion. Contributing to micropexophagy, PMN. Forms the nucleus-vacuole junction by binding with nuclear envelope protein Nvj1 during PMN. | [18,19] | N.I. |
| Niemann–Pick type C proteins (NPC) | Ncr1, Npc2 | Form membrane rafts on the vacuolar membrane by transporting sterol during microlipophagy both in the stationary phase and in acute nitrogen starvation in yeast. | [23] | NPC1 [45] |
| VTC complex | Vtc1, Vtc2, Vtc3, Vtc4 | Involved in the tubule formation on the vacuolar membrane. Recruited on the vacuole in nitrogen starvation. Directly binds to calmodulin in microautophagy in yeast. | [46] | N.I. |
| ESCRT complex and related proteins | ESCRT-0: Hse1, Bro1, Vps27 ESCRT-I: Vps23, Vps28, Vps3, Mvb12 ESCRT-II: Vps22, Vps25, Vps36 ESCRT-III: Vps2, Vps20, Vps24, Snf7, etc. VPS4-VTA1: Vps4 | Expected to contribute to membrane bending, remodeling, and scission during micro-ER-phagy in yeast and endosomal microautophagy in animals. Full activity of Vps27 (binding to ubiquitin, PI3P, and ESCRT-I) is required for diauxic shift-induced lipophagy (yeast). Formation of lipid domains on the vacuolar membrane in response to nutrient deprivation (yeast). | [14,29,30,47] | ESCRT-I to -III, VPS4–VTA1, functional analogue of ESCRT-0, plant-specific ESCRT [48,49,50,51] |
| Nem1–Spo7 complex | Nem1, Spo7, Pah1 | Expected to contribute to membrane remodeling with ESCRTs during micro-ER-phagy in yeast. | [29] | PAH1,2 [52] |
| Clathrin | Chc1 | Interacts with Vps27 (ESCRT-0). Required for microlipophagy in the diauxic shift (yeast). | [30] | CHC1, CHC2 [53] |
| Vacuole fusion | SNARE: Vam3, Vam7 SNAP: Sec17 NSF: Sec18 HOPS: Vps18, Vps33, Vps39, Vps41 | Can be involved in membrane fusion processes. Required for PMN and micro-ER-phagy. | [19,30] | SNARE and HOPS complexes, including α-SNAP/SEC17, NSF/SEC18, SYP22/VAM3, VPS18, 33, 39, 41 [54,55,56,57] |
| Selective receptor of autophagy | Nbr1 | Selective receptor of cytosolic ubiquitinated cargo in endosomal microautophagy. | [58] | NBR1 [59] |
| Hsc70 | Selective receptor of cytosolic proteins in endosomal microautophagy. | [14,60] | Cytosolic/nuclear HSC70-1 to -5 [61,62] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sieńko, K.; Poormassalehgoo, A.; Yamada, K.; Goto-Yamada, S. Microautophagy in Plants: Consideration of Its Molecular Mechanism. Cells 2020, 9, 887. https://doi.org/10.3390/cells9040887
Sieńko K, Poormassalehgoo A, Yamada K, Goto-Yamada S. Microautophagy in Plants: Consideration of Its Molecular Mechanism. Cells. 2020; 9(4):887. https://doi.org/10.3390/cells9040887
Chicago/Turabian StyleSieńko, Katarzyna, Andisheh Poormassalehgoo, Kenji Yamada, and Shino Goto-Yamada. 2020. "Microautophagy in Plants: Consideration of Its Molecular Mechanism" Cells 9, no. 4: 887. https://doi.org/10.3390/cells9040887
APA StyleSieńko, K., Poormassalehgoo, A., Yamada, K., & Goto-Yamada, S. (2020). Microautophagy in Plants: Consideration of Its Molecular Mechanism. Cells, 9(4), 887. https://doi.org/10.3390/cells9040887