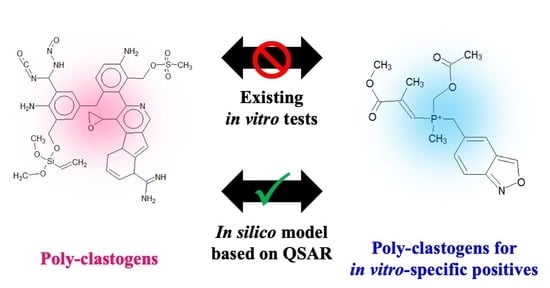
In Silico Model for Chemical-Induced Chromosomal Damages Elucidates Mode of Action and Irrelevant Positives
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data set of Chemical Substances
2.1.1. Data Acquisition and Classification
- Positives: (a) chemicals with “positive results in in vitro mammalian cell genotoxicity tests” in Kirkland et al. [12] (25 chemicals) and (b) positive chemicals (70 chemicals) and “chemicals with minimal or some concern” (12 chemicals) in Morita et al [11]. Although o-Dichlorobenzene (CAS No. 95-50-1) had been classified into “missed chemicals with negligible concern” in Morita et al. [11], it was recategorized into positives because positive results were recently reported in both in vivo [20] and in vitro [15] micronucleus tests under the current OECD test guidelines [14]. In total, 108 chemicals were classified as positives.
- Misleading positives: (a) chemicals that “should give negative results in in vitro mammalian cell genotoxicity tests, but have been reported to induce gene mutations in mouse lymphoma cells, chromosomal aberrations, or micronuclei, often at high concentrations or at high levels of cytotoxicity” in Kirkland et al. [12] (17 chemicals), (b) “chemicals with negligible concern” (25 chemicals) in Morita et al. [11], and (c) among chemicals with negative Ames tests in Morita et al. [11], chemicals that were suggested to be misleading positives owing to cytotoxicity [16] and showed negative retest results using in vitro micronucleus test in Fujita et al. [15] (8 chemicals). In total, 50 chemicals were classified as misleading positives. Basically, misleading positive chemicals do not induce genotoxicity in in vivo conditions and induce irreverent positives in in vitro conditions.
2.1.2. Reselection of Chemicals via OFG Extraction
2.2. Prediction Model Development
2.3. Performance Evaluation of Models
2.4. Visualization of Structural Alerts (OFGs)
3. Results
3.1. Prediction Performances of Developed Model
3.2. OFGs Related to Test Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Environment Report of the Expert Consultation on Scientific and Regulatory Evaluation of Organic Chemistry Mechanism-Based Structural Alerts for the Identification of DNA Binding Chemicals PART2; OECD: Paris, France, 2010.
- Tcheremenskaia, O.; Battistelli, C.L.; Giuliani, A.; Benigni, R.; Bossa, C. In silico approaches for prediction of genotoxic and carcinogenic potential of cosmetic ingredients. Comput. Toxicol. 2019, 11, 91–100. [Google Scholar] [CrossRef]
- European Union Directive 2003/15/EC of the European Parliament and of the Council of 27 February 2003 amending Council Directive 76/768/EEC on the approximation of the laws of the Member States relating to cosmetic products. Off. J. Eur. Union 2003, 66, 26–35.
- ICH Guideline S2 (R1) on Genotoxicity Testing and Data Interpretation for Pharmaceuticals Intended for Human Use; European Medicines Agency: London, UK, 2008; pp. 1–28.
- Kirkland, D.; Aardema, M.; Henderson, L.; Müller, L. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens: I. Sensitivity, specificity and relative predictivity. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2005, 584, 1–256. [Google Scholar] [CrossRef] [PubMed]
- ICH Guideline M7(R1) on Assessment and Control of DNA Reactive (Mutagenic) Impurities in Pharmaceuticals to Limit Potential Carcinogenic Risk; European Medicines Agency: London, UK, 2015; pp. 1–110.
- Morita, T.; Shigeta, Y.; Kawamura, T.; Fujita, Y.; Honda, H.; Honma, M. In silico prediction of chromosome damage: Comparison of three (Q) SAR models. Mutagenesis 2019, 34, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, D.; Aardema, M.; Müller, L.; Hayashi, M. Evaluation of the ability of a battery of three in vitro genotoxicity tests to discriminate rodent carcinogens and non-carcinogens. II. Further analysis of mammalian cell results, relative predictivity and tumour profiles. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2006, 608, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Rothfuss, A.; Steger-Hartmann, T.; Heinrich, N.; Wichard, J. Computational prediction of the chromosome-damaging potential of chemicals. Chem. Res. Toxicol. 2006, 19, 1313–1319. [Google Scholar] [CrossRef]
- Honda, H.; Fujita, Y.; Kasamatsu, T.; Fuchs, A.; Fautz, R.; Morita, O. Necessity for retrospective evaluation of past-positive chemicals in in vitro chromosomal aberration tests using recommended cytotoxicity indices. Genes Environ. 2018, 40, 2. [Google Scholar] [CrossRef] [Green Version]
- Morita, T.; Honma, M.; Morikawa, K. Effect of reducing the top concentration used in the in vitro chromosomal aberration test in CHL cells on the evaluation of industrial chemical genotoxicity. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2012, 741, 32–56. [Google Scholar] [CrossRef]
- Kirkland, D.; Kasper, P.; Martus, H.J.; Müller, L.; van Benthem, J.; Madia, F.; Corvi, R. Updated recommended lists of genotoxic and non-genotoxic chemicals for assessment of the performance of new or improved genotoxicity tests. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 795, 7–30. [Google Scholar] [CrossRef]
- OECD OECD TG473. In vitro mammalian chromosomal aberration test. In Test Guidelines for the Chemicals; OECD: Paris, French, 2014; pp. 1–20. [Google Scholar] [CrossRef]
- OECD OECD TG487. In vitro mammalian cell micronucleus test. In Test Guidelines for the Chemicals; OECD: Paris, French, 2014; pp. 1–26. [Google Scholar] [CrossRef]
- Fujita, Y.; Ito, Y.; Morita, O.; Honda, H. Validation of retrospective evaluation method for false genotoxic chemicals with strong cytotoxicity: Re-evaluation using in vitro micronucleus test. Fundam. Toxicol. Sci. 2016, 3, 251–256. [Google Scholar] [CrossRef]
- Fujita, Y.; Morita, T.; Matsumura, S.; Kawamoto, T.; Ito, Y.; Nishiyama, N.; Honda, H. Comprehensive retrospective evaluation of existing in vitro chromosomal aberration test data by cytotoxicity index transformation. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 802, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Kawamoto, T.; Morita, O.; Yoshinari, K.; Honda, H. Discriminating between adaptive and carcinogenic liver hypertrophy in rat studies using logistic ridge regression analysis of toxicogenomic data: The mode of action and predictive models. Toxicol. Appl. Pharmacol. 2017, 318, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Amano, Y.; Honda, H.; Sawada, R.; Nukada, Y.; Yamane, M.; Ikeda, N.; Morita, O.; Yamanishi, Y. In silico systems for predicting chemical-induced side effects using known and potential chemical protein interactions, enabling mechanism estimation. J. Toxicol. Sci. 2020, 45, 137–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Kasamatsu, T.; Ikeda, N.; Nishiyama, N.; Honda, H. A retrospective evaluation method for in vitro mammalian genotoxicity tests using cytotoxicity index transformation formulae. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 796, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Toxicological Review of Dichlorobenzenes; Environmental Protection Agency: Washington, DC, USA, 2006.
- OECD. The OECD QSAR Toolbox. Available online: http://www.oecd.org/chemicalsafety/risk-assessment/theoecdqsartoolbox.htm (accessed on 9 December 2016).
- Friedman, J.; Hastie, T.; Simon, N.; Tibshirani, R. Package ‘Glmnet’. Available online: https://cran.r-project.org/web/packages/glmnet/index.html (accessed on 9 December 2016).
- Hastie, T.; Qian, J. Glmnet vignette. Retriev. June 2016, 2014, 1–42. [Google Scholar]
- Szumilas, M. Explaining odds ratios. J. Can. Acad. Child Adolesc. Psychiatry 2010, 19, 227–229. [Google Scholar] [PubMed]
- Dearden, J.C.; Cronin, M.T.D.; Kaiser, K.L.E. How not to develop a quantitative structure-activity or structure-property relationship (QSAR/QSPR). SAR QSAR Environ. Res. 2009, 20, 241–266. [Google Scholar] [CrossRef] [PubMed]
- Torgo, L. Package ‘DMwR’. Available online: https://cran.r-project.org/web/packages/DMwR/DMwR.pdf (accessed on 9 December 2016).
- Takeshita, J.; Nakayama, H.; Kitsunai, Y.; Tanabe, M.; Oki, H.; Sasaki, T.; Yoshinari, K. Discriminative models using molecular descriptors for predicting increased serum ALT levels in repeated-dose toxicity studies of rats. Comput. Toxicol. 2017, 6, 64–70. [Google Scholar] [CrossRef]
- Engebretsen, S.; Bohlin, J. Statistical predictions with glmnet. Clin. Epigenetics 2019, 11, 123. [Google Scholar] [CrossRef] [Green Version]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Beleites, C.; Salzer, R.; Sergo, V. Validation of soft classification models using partial class memberships: An extended concept of sensitivity & co. applied to grading of astrocytoma tissues. Chemom. Intell. Lab. Syst. 2013, 122, 12–22. [Google Scholar] [CrossRef] [Green Version]
- Ashby, J.; Tennant, R.W. Chemical structure, Salmonella mutagenicity and extent of carcinogenicity as indicators of genotoxic carcinogenesis among 222 chemicals tested in rodents by the U.S. NCI/NTP. Mutat. Res. 1988, 204, 17–115. [Google Scholar] [CrossRef]
- Ashby, J.; Tennant, R.W. Definitive relationships among chemical structure, carcinogenicity and mutagenicity for 301 chemicals tested by the U.S. NTP. Mutat. Res. Genet. Toxicol. 1991, 257, 229–306. [Google Scholar] [CrossRef]
- Gonzales, G.B.; de Saeger, S. Elastic net regularized regression for time-series analysis of plasma metabolome stability under sub-optimal freezing condition. Sci. Rep. 2018, 8, 3659. [Google Scholar] [CrossRef] [Green Version]
- Mehrmohamadi, M.; Mentch, L.K.; Clark, A.G.; Locasale, J.W. Integrative modelling of tumour DNA methylation quantifies the contribution of metabolism. Nat. Commun. 2016, 7, 13666. [Google Scholar] [CrossRef] [Green Version]
- Environment Report of the Expert Consultation on Scientific and Regulatory Evaluation of Organic Chemistry Mechanism-Based Structural Alerts for the Identification of DNA Binding Chemicals PART1; OECD: Paris, France, 2010; Volume 120.
- Nguyen, B.; Hamelberg, D.; Bailly, C.; Colson, P.; Stanek, J.; Brun, R.; Neidle, S.; Wilson, W.D. Characterization of a novel DNA minor-groove complex. Biophys. J. 2004, 86, 1028–1041. [Google Scholar] [CrossRef] [Green Version]
- Moraski, G.C.; Chang, M.; Villegas-Estrada, A.; Franzblau, S.G.; Möllmann, U.; Miller, M.J. Structure-activity relationship of new anti-tuberculosis agents derived from oxazoline and oxazole benzyl esters. Eur. J. Med. Chem. 2010, 45, 1703–1716. [Google Scholar] [CrossRef] [Green Version]
- Gerner, I.; Barratt, M.D.; Zinke, S.; Schlegel, K.; Schlede, E. Development and prevalidation of a list of structure-activity relationship rules to be used in expert systems for prediction of the skin-sensitising properties of chemicals. ATLA Altern. Lab. Anim. 2004, 32, 487–509. [Google Scholar] [CrossRef]
- Razzaque, M.S. Phosphate toxicity: New insights into an old problem. Clin. Sci. 2012, 120, 91–97. [Google Scholar] [CrossRef] [Green Version]
- Corvi, R.; Albertini, S.; Hartung, T.; Hoffmann, S.; Maurici, D.; Pfuhler, S.; van Benthem, J.; Vanparys, P. ECVAM retrospective validation of in vitro micronucleus test (MNT). Mutagenesis 2008, 23, 271–283. [Google Scholar] [CrossRef]
- Fowler, P.; Smith, R.; Smith, K.; Young, J.; Jeffrey, L.; Carmichael, P.; Kirkland, D.; Pfuhler, S. Reduction of misleading (“false”) positive results in mammalian cell genotoxicity assays. III: Sensitivity of human cell types to known genotoxic agents. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014, 767, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Snyder, R.D.; Pearl, G.S.; Mandakas, G.; Choy, W.N.; Goodsaid, F.; Rosenblum, I.Y. Assessment of the sensitivity of the computational programs DEREK, TOPKAT, and MCASE in the prediction of the genotoxicity of pharmaceutical molecules. Environ. Mol. Mutagen. 2004, 43, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, D.; Zeiger, E.; Madia, F.; Gooderham, N.; Kasper, P.; Lynch, A.; Morita, T.; Ouedraogo, G.; Parra Morte, J.M.; Pfuhler, S.; et al. Can in vitro mammalian cell genotoxicity test results be used to complement positive results in the Ames test and help predict carcinogenic or in vivo genotoxic activity? I. Reports of individual databases presented at an EURL ECVAM Workshop. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014, 775, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Canipa, S.; Cayley, A.; Drewe, W.C.; Williams, R.V.; Hamada, S.; Hirose, A.; Honma, M.; Morita, T. Using in vitro structural alerts for chromosome damage to predict in vivo activity and direct future testing. Mutagenesis 2016, 31, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Estrada, E.; Molina, E. Automatic extraction of structural alerts for predicting chromosome aberrations of organic compounds. J. Mol. Graph. Model. 2006, 25, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Kulkarni, R.; Hewitt, N.J.; Aardema, M.J. The EpiDermTM 3D human reconstructed skin micronucleus (RSMN) assay: Historical control data and proof of principle studies for mechanistic assay adaptations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 805, 25–37. [Google Scholar] [CrossRef]
- Pfuhler, S.; Reisinger, K. Reconstructed skin micronucleus assay (RSMN). In Alternatives for Dermal Toxicity Testing; Springer International Publishing: Berlin, Germany, 2017; pp. 513–525. ISBN 9783319503530. [Google Scholar]
- Kirkland, D.J.; Aardema, M.; Banduhn, N.; Carmichael, P.; Fautz, R.; Meunier, J.R.; Pfuhler, S. In vitro approaches to develop weight of evidence (WoE) and mode of action (MoA) discussions with positive in vitro genotoxicity results. Mutagenesis 2007, 22, 161–175. [Google Scholar] [CrossRef] [Green Version]
- Fowler, P.; Smith, K.; Young, J.; Jeffrey, L.; Kirkland, D.; Pfuhler, S.; Carmichael, P. Reduction of misleading (“false”) positive results in mammalian cell genotoxicity assays. I. Choice of cell type. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2012, 742, 11–25. [Google Scholar] [CrossRef]
- Fujita, Y.; Honda, H.; Yamane, M.; Morita, T.; Matsuda, T.; Morita, O. A decision tree-based integrated testing strategy for tailor-made carcinogenicity evaluation of test substances using genotoxicity test results and chemical spaces. Mutagenesis 2019, 34, 3–16. [Google Scholar] [CrossRef]


| Accuracy (%) | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|
| P | 85.6 | 72.6 | 92.7 |
| N | 80.3 | 71.0 | 85.2 |
| MP | 87.9 | 71.6 | 94.8 |
| OFG | Odds Ratio | Reported Main Toxicological Effect or Mechanisms Likely Related to Positive Results | REF |
|---|---|---|---|
| Epoxide | 13.94 | DNA binding (a) | [35] |
| Fused unsaturated carbocycles | 10.84 | metabolites: DNA binding (c) * | [21] |
| Alkoxysilane | 10.21 | DNA binding (a) | [11] |
| Sulfonate ester | 9.16 | DNA binding (a) | [35] |
| Fused heterocyclic aromatic | 9.14 | DNA intercalation (c) | [35] |
| N. Nitroso | 9.09 | DNA binding (a) | [35] |
| Amidine | 8.34 | DNA minor groove binders (b) | [36] |
| Isocyanate | 8.34 | DNA acylation (a) | [35] |
| Dianilines | 8.28 | DNA binding (c) | [21] |
| OFG | Odds Ratio | Reported Main Toxicological Effects or Mechanisms Likely Related to Misleading Positive Results | REF |
|---|---|---|---|
| Oxazole/Izoxazole | 12.32 | Anti-tuberculosis activity (a) | [37] |
| Benzthiazolinone/Benzoisothiazolinone | 11.83 | Reaction with amino groups of lysine residues (b) | [38] |
| Phosphonium, salt | 7.68 | Cytotoxicity (b) | [39] |
| Acetoxy | 4.09 | Low pH (a) | [11] |
| Methacrylate | 4.05 | DNA reactivity in vitro-specific and/or cytotoxicity (b) | [11] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujita, Y.; Morita, O.; Honda, H. In Silico Model for Chemical-Induced Chromosomal Damages Elucidates Mode of Action and Irrelevant Positives. Genes 2020, 11, 1181. https://doi.org/10.3390/genes11101181
Fujita Y, Morita O, Honda H. In Silico Model for Chemical-Induced Chromosomal Damages Elucidates Mode of Action and Irrelevant Positives. Genes. 2020; 11(10):1181. https://doi.org/10.3390/genes11101181
Chicago/Turabian StyleFujita, Yurika, Osamu Morita, and Hiroshi Honda. 2020. "In Silico Model for Chemical-Induced Chromosomal Damages Elucidates Mode of Action and Irrelevant Positives" Genes 11, no. 10: 1181. https://doi.org/10.3390/genes11101181
APA StyleFujita, Y., Morita, O., & Honda, H. (2020). In Silico Model for Chemical-Induced Chromosomal Damages Elucidates Mode of Action and Irrelevant Positives. Genes, 11(10), 1181. https://doi.org/10.3390/genes11101181