Insights into the Genetic Architecture of Bran Friability and Water Retention Capacity, Two Important Traits for Whole Grain End-Use Quality in Winter Wheat
Abstract
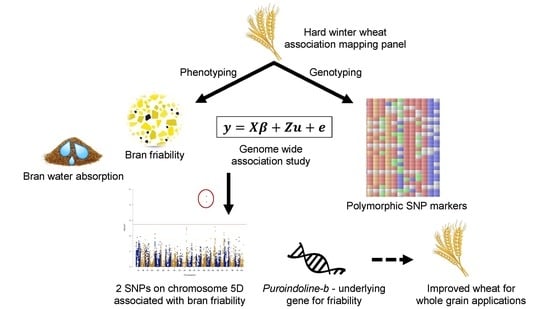
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Bran Functionality Traits
2.3. Statistical Analysis of Bran Functionality Traits
2.4. Genotyping and Population Structure
2.5. Estimating Heritability Using Genome-Wide SNP Markers
2.6. Genome-Wide Association Study (GWAS)
2.7. Candidate Gene Analysis
2.8. Linkage Disequilibrium between Trait-Associated SNP Markers on Chromosome 5D and Pin Locus
3. Results and Discussion
3.1. Bran Functionality Traits
3.2. Genotyping and Population Structure
3.3. Heritability Estimates for Bran Functionality Traits Using Genome-Wide SNP Markers
3.4. GWAS for Bran Friability
3.5. GWAS for Bran WRC
3.6. Candidate Genomic Regions for Bran Friability and WRC
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aune, D.; Norat, T.; Romundstad, P.; Vatten, L.J. Whole grain and refined grain consumption and the risk of type 2 diabetes: A systematic review and dose-response meta-analysis of cohort studies. Eur. J. Epidemiol. 2013, 28, 845–858. [Google Scholar] [CrossRef]
- Cho, S.S.; Qi, L.; Fahey, G.C.J.; Klurfeld, D.M. Consumption of cereal fiber, mixtures of whole grains and bran, and whole grains and risk reduction in type 2 diabetes, obesity, and cardiovascular disease. Am. J. Clin. Nutr. 2013, 98, 594–619. [Google Scholar] [CrossRef] [PubMed]
- Whole Grains Council. Whole Grain Statistics [Internet]. 2020. Available online: https://wholegrainscouncil.org/newsroom/whole-grain-statistics (accessed on 13 March 2020).
- Whole Grain Initiative. Definition of Whole Grain as Food Ingredient [Internet]. 2019. Available online: https://wgi.meetinghand.com/projectData/775/webData/Definition-of-Whole-Grain-as-Food-Ingredient-Version-20190501C.pdf (accessed on 8 July 2020).
- Cai, L.; Choi, I.; Lee, C.K.; Park, K.K.; Baik, B.K. Bran characteristics and bread-baking quality of whole grain wheat flour. Cereal Chem. 2014, 91, 398–405. [Google Scholar] [CrossRef]
- Noort, M.W.J.; van Haaster, D.; Hemery, Y.; Schols, H.A.; Hamer, R.A. The effect of particle size of wheat bran fractions on bread quality-Evidence for fibre-protein interactions. J. Cereal Sci. 2010, 52, 59–64. [Google Scholar] [CrossRef]
- Jacobs, P.J.; Hemdane, S.; Claes, S.; Mulders, L.; Langenaeken, N.A.; Dewettinck, K.; Courtin, C.M. Wheat bran-associated subaleurone and endosperm proteins and their impact on bran-rich bread-making. J. Cereal Sci. 2018, 81, 99–107. [Google Scholar] [CrossRef]
- Jacobs, P.J.; Hemdane, S.; Dornez, E.; Delcour, J.A.; Courtin, C.M. Study of hydration properties of wheat bran as a function of particle size. Food Chem. 2015, 179, 296–304. [Google Scholar] [CrossRef]
- Navrotskyi, S.; Guo, G.; Baenziger, P.S.; Xu, L.; Rose, D.J. Impact of wheat bran physical properties and chemical composition on whole grain flour mixing and baking properties. J. Cereal Sci. 2019, 89, 102790. [Google Scholar] [CrossRef]
- Khalid, K.H.; Manthey, F.; Simsek, S. Centrifugal milling of wheat bran. Cereal Chem. 2018, 95, 330–341. [Google Scholar] [CrossRef]
- Zhang, D.; Moore, W.R. Effect of wheat bran particle size on dough rheological properties. J. Sci. Food Agric. 1997, 74, 490–496. [Google Scholar] [CrossRef]
- Hemdane, S.; Jacobs, P.J.; Dornez, E.; Verspreet, J.; Delcour, J.A.; Courtin, C.M. Wheat (Triticum aestivum L.) bran in bread making: A critical review. Compr. Rev. Food Sci. Food Saf. 2016, 15, 28–42. [Google Scholar] [CrossRef] [Green Version]
- Maccaferri, M.; Zhang, J.; Bulli, P.; Abate, Z.; Chao, S.; Cantu, D.; Bossolini, E.; Chen, X.; Pumphrey, M.; Dubcovsky, J. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3 Genes Genomes Genet. 2015, 5, 449–465. [Google Scholar]
- Jaiswal, V.; Gahlaut, V.; Meher, P.K.; Mir, R.R.; Jaiswal, J.P.; Rao, A.; Balyan, H.S.; Gupta, P. Genome Wide Single Locus Single Trait, Multi-Locus and Multi-Trait Association Mapping for Some Important Agronomic Traits in Common Wheat (T. aestivum L.). PLoS ONE 2016, 11, e0159343. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, H.; Serfling, A.; Friedt, W.; Ordon, F. Genome-Wide Association Studies Reveal Genomic Regions Associated With the Response of Wheat (Triticum aestivum L.) to Mycorrhizae Under Drought Stress Conditions. Front. Plant Sci. 2018, 9, 1728. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhao, L.; Ren, Y.; Dong, Z.; Cui, D.; Chen, F. Genome-wide association study revealed that the TaGW8 gene was associated with kernel size in Chinese bread wheat. Sci. Rep. 2019, 9, 2702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamil, M.; Ali, A.; Gul, A.; Ghafoor, A.; Napar, A.A.; Ibrahim, A.M.H.; Naveed, N.H.; Yasin, N.A.; Mujeeb-Kazi, A. Genome-wide association studies of seven agronomic traits under two sowing conditions in bread wheat. BMC Plant Biol. 2019, 19, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Pan, Y.; Singh, P.K.; He, X.; Ren, Y.; Zhao, L.; Zhang, N.; Cheng, S.; Chen, F. Investigation and genome-wide association study for Fusarium crown rot resistance in Chinese common wheat. BMC Plant Biol. 2019, 19, 153. [Google Scholar] [CrossRef] [Green Version]
- Sheoran, S.; Jaiswal, S.; Kumar, D.; Raghav, N.; Sharma, R.; Pawar, S.; Paul, S.; Iquebal, M.A.; Jaiswar, A.; Sharma, P.; et al. Uncovering Genomic Regions Associated With 36 Agro-Morphological Traits in Indian Spring Wheat Using GWAS. Front. Plant Sci. 2019, 10, 527. [Google Scholar] [CrossRef]
- Afzal, F.; Li, H.; Gul, A.; Subhani, A.; Ali, A.; Mujeeb-Kazi, A.; Ogbonnaya, F.; Trethowan, R.; Xia, X.; He, Z.; et al. Genome-Wide Analyses Reveal Footprints of Divergent Selection and Drought Adaptive Traits in Synthetic-Derived Wheats. G3 Genes Genomes Genet. 2019, 9, 1957–1973. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.; Babar, A.; Robbins, K.; Bai, G.; Mason, R.E.; Khan, J.; Shahi, D.; Avci, M.; Guo, J.; Hossain, M.M.; et al. Understanding the Genetic Basis of Spike Fertility to Improve Grain Number, Harvest Index, and Grain Yield in Wheat Under High Temperature Stress Environments. Front. Plant Sci. 2019, 10, 1481. [Google Scholar] [CrossRef] [Green Version]
- Rahimi, Y.; Bihamta, M.R.; Taleei, A.; Alipour, H.; Ingvarsson, P.K. Genome-wide association study of agronomic traits in bread wheat reveals novel putative alleles for future breeding programs. BMC Plant Biol. 2019, 19, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Basile, S.M.L.; Ramírez, I.A.; Crescente, J.M.; Conde, M.B.; Demichelis, M.; Abbate, P.; Rogers, W.J.; Pontaroli, A.; Helguera, M.; Vanzetti, L.S. Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biol. 2019, 19, 1–16. [Google Scholar]
- Bin Safdar, L.; Andleeb, T.; Latif, S.; Umer, M.J.; Tang, M.; Li, X.; Liu, S.; Quraishi, U.M. Genome-Wide Association Study and QTL Meta-Analysis Identified Novel Genomic Loci Controlling Potassium Use Efficiency and Agronomic Traits in Bread Wheat. Front. Plant Sci. 2020, 11, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Xu, X.; Bai, G.; Carver, B.F.; Hunger, R.; Bonman, J.M.; Kolmer, J.A.; Dong, H. Genome-Wide Association Mapping Reveals Novel QTL for Seedling Leaf Rust Resistance in a Worldwide Collection of Winter Wheat. Plant Genome 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daba, S.D.; Tyagi, P.; Brown-Guedira, G.; Mohammadi, M. Genome-wide association study in historical and contemporary U.S. winter wheats identifies height-reducing loci. Crop J. 2020, 8, 243–251. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, F.; Yan, X.; Zhang, X.; Dong, Z.; Cui, D.; Chen, F. Genome-wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant Biotechnol. J. 2017, 15, 953–969. [Google Scholar] [CrossRef] [Green Version]
- Ogbonnaya, F.C.; Rasheed, A.; Okechukwu, E.C.; Jighly, A.; Makdis, F.; Wuletaw, T.; Hagras, A.; Uguru, M.I.; Agbo, C.U. Genome-wide association study for agronomic and physiological traits in spring wheat evaluated in a range of heat prone environments. Theor. Appl. Genet. 2017, 130, 1819–1835. [Google Scholar] [CrossRef]
- Liu, J.; He, Z.; Rasheed, A.; Wen, W.; Yan, J.; Zhang, P.; Wan, Y.; Zhang, Y.; Xie, C.; Xia, X. Genome-wide association mapping of black point reaction in common wheat (Triticum aestivum L.). BMC Plant Biol. 2017, 17, 220. [Google Scholar] [CrossRef] [Green Version]
- Sukumaran, S.; Lopes, M.; Dreisigacker, S.; Reynolds, M. Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor. Appl. Genet. 2018, 131, 985–998. [Google Scholar] [CrossRef]
- Peng, Y.; Liu, H.; Chen, J.; Shi, T.; Zhang, C.; Sun, D.; He, Z.; Hao, Y.; Chen, W. Genome-Wide Association Studies of Free Amino Acid Levels by Six Multi-Locus Models in Bread Wheat. Front. Plant Sci. 2018, 9, 1196. [Google Scholar]
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Baenziger, P.S. Genome-Wide Association Study Reveals Novel Genomic Regions for Grain Yield and Yield-Related Traits in Drought-Stressed Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018, 19, 3011. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Xu, Z.; Fan, X.; Zhou, Q.; Cao, J.; Wang, F.; Ji, G.; Yang, L.; Feng, B.; Wang, T. A Genome-Wide Association Study of Wheat Spike Related Traits in China. Front. Plant Sci. 2018, 9, 1584. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Shamanin, V.; Shepelev, S.S.; Baenziger, P.S.; Pozherukova, V.; Pototskaya, I.; Morgounov, A. Marker-Trait Associations for Enhancing Agronomic Performance, Disease Resistance, and Grain Quality in Synthetic and Bread Wheat Accessions in Western Siberia. G3 Genes Genomes Genet. 2019, 9, 4209–4222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Wang, S.; Wei, W.; Xie, H.; Liu, K.; Zhang, C.; Wu, Z.; Jiang, H.; Cao, J.; Zhao, L.; et al. Genome-wide association study of pre-harvest sprouting tolerance using a 90K SNP array in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2019, 132, 2947–2963. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, F.; Zhao, C.; Lv, G.; Sun, C.; Pan, Y.; Guo, X.; Chen, F. Genome-wide association study of six quality traits reveals the association of the TaRPP13L1 gene with flour colour in Chinese bread wheat. Plant Biotechnol. J. 2019, 17, 2106–2122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, P.S.; Jensen, J.; Andersen, J.R.; Guzmán, C.; Orabi, J.; Jahoor, A. Genomic Prediction and Genome-Wide Association Studies of Flour Yield and Alveograph Quality Traits Using Advanced Winter Wheat Breeding Material. Genes 2019, 10, 669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battenfield, S.D.; Sheridan, J.L.; Silva, L.D.C.E.; Miclaus, K.J.; Dreisigacker, S.; Wolfinger, R.D.; Peña, R.J.; Singh, R.P.; Jackson, E.W.; Fritz, A.K.; et al. Breeding-assisted genomics: Applying meta-GWAS for milling and baking quality in CIMMYT wheat breeding program. PLoS ONE 2018, 13, e0204757. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.; Liu, J.; Xu, D.; Wen, W.; Yan, J.; Zhang, P.; Wan, Y.; Cao, S.; Hao, Y.; Xia, X.; et al. A Genome-Wide Association Study Reveals a Rich Genetic Architecture of Flour Color-Related Traits in Bread Wheat. Front. Plant Sci. 2018, 9, 1136. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Feng, B.; Xu, Z.; Fan, X.; Jiang, F.; Jin, X.; Cao, J.; Wang, F.; Liu, Q.; Yang, L.; et al. A genome-wide association study of wheat yield and quality-related traits in southwest China. Mol. Breed. 2017, 38, 1–11. [Google Scholar] [CrossRef]
- Li, J.; Rasheed, A.; Guo, Q.; Dong, Y.; Liu, J.; Xia, X.; Zhang, Y.; He, Z. Genome-wide association mapping of starch granule size distribution in common wheat. J. Cereal Sci. 2017, 77, 211–218. [Google Scholar] [CrossRef]
- Grogan, S.M.; Anderson, J.; Baenziger, P.S.; Frels, K.; Guttieri, M.J.; Haley, S.D.; Kim, K.-S.; Liu, S.; McMaster, G.S.; Newell, M.; et al. Phenotypic Plasticity of Winter Wheat Heading Date and Grain Yield across the US Great Plains. Crop Sci. 2016, 56, 2223–2236. [Google Scholar] [CrossRef]
- Guttieri, M.J.; Baenziger, P.S.; Frels, K.; Carver, B.; Arnall, B.; Waters, B. Variation for Grain Mineral Concentration in a Diversity Panel of Current and Historical Great Plains Hard Winter Wheat Germplasm. Crop Sci. 2015, 55, 1035–1052. [Google Scholar] [CrossRef]
- Guttieri, M.J.; Baenziger, P.S.; Frels, K.; Carver, B.; Arnall, B.; Wang, S.; Akhunov, E.; Waters, B. Prospects for Selecting Wheat with Increased Zinc and Decreased Cadmium Concentration in Grain. Crop Sci. 2015, 55, 1712–1728. [Google Scholar] [CrossRef]
- Ayana, G.T.; Ali, S.; Sidhu, J.S.; Hernandez, J.L.G.; Turnipseed, B.; Sehgal, S.K. Genome-Wide Association Study for Spot Blotch Resistance in Hard Winter Wheat. Front. Plant Sci. 2018, 9, 926. [Google Scholar]
- Awad, W. Genome-Wide Association Study and Drought Tolerance Evaluation of a Winter Wheat Association Mapping Panel. Ph.D. Thesis, Colorado State University, Fort Collins, CO, USA, 2015. [Google Scholar]
- Bhatta, M.; Baenziger, P.S.; Waters, B.; Poudel, R.; Belamkar, V.; Poland, J.; Morgounov, A. Genome-Wide Association Study Reveals Novel Genomic Regions Associated with 10 Grain Minerals in Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2018, 19, 3237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, S.M.; Sidhu, J.S.; Ali, S.; Kaur, N.; Wu, J.; Sehgal, S.K. Molecular characterization of bacterial leaf streak resistance in hard winter wheat. PeerJ 2019, 7, e7276. [Google Scholar] [CrossRef] [Green Version]
- Cereals & Grains Association. Approved methods: 26-50.01–Brabender Quadrumat Jr. (Quadruplex) method; 56-11.02–Solvent retention capacity profile. In Cereals & Grains Association, editor. Approv. Methods Anal, 11th ed.; Cereals & Grains Association: St. Louis, MO, USA, 2020; Available online: http://methods.aaccnet.org/default.aspx (accessed on 22 July 2020).
- T3/Wheat [Internet]. Available online: https://triticeaetoolbox.org/wheat/genotyping/display_genotype.php?trial_code=TCAP90K_HWWAMP (accessed on 22 July 2020).
- El-Basyoni, I.S.; Lorenz, A.; Guttieri, M.; Frels, K.; Baenziger, P.; Poland, J.; Akhunov, E. A comparison between genotyping-by-sequencing and array-based scoring of SNPs for genomic prediction accuracy in winter wheat. Plant Sci. 2018, 270, 123–130. [Google Scholar] [CrossRef]
- The International Wheat Genome Sequencing Consortium (IWGSC); Appels, R.; Eversole, K.A.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar719. [Google Scholar]
- Belamkar, V.; Farmer, A.D.; Weeks, N.T.; Kalberer, S.R.; Blackmon, W.J.; Cannon, S.B. Genomics-assisted characterization of a breeding collection of Apios americana, an edible tuberous legume. Sci. Rep. 2016, 6, 34908. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Battenfield, S.D.; Guzman, C.; Gaynor, R.C.; Singh, R.P.; Peña, R.J.; Dreisigacker, S.; Fritz, A.K.; Poland, J. Genomic Selection for Processing and End-Use Quality Traits in the CIMMYT Spring Bread Wheat Breeding Program. Plant Genome 2016, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez, P.; Campos, G.D.L. Genome-Wide Regression and Prediction with the BGLR Statistical Package. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Belamkar, V.; Guttieri, M.J.; Hussain, W.; Jarquín, D.; El-Basyoni, I.; Poland, J.; Lorenz, A.J.; Baenziger, P.S. Genomic Selection in Preliminary Yield Trials in a Winter Wheat Breeding Program. G3 Genes Genomes Genet. 2018, 8, 2735–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oraguzie, N.; Rikkerink, E.; Gardine, S.; De Silva, N. Association Mapping in Plants; Oraguzie, N., Rikkerink, E., Gardine, S., De Silva, N., Eds.; Springer: New York, NY, USA, 2007. [Google Scholar]
- Collins, A. Linkage Disequilibrium and Association Mapping: Analysis and Applications; Collins, A., Ed.; Humana Press: Totowa, NJ, USA, 2007. [Google Scholar]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 005165. [Google Scholar] [CrossRef] [Green Version]
- U.S. Department of Health and Human Services, U.S. Department of Agriculture. Dietary Guidelines for Americans 2015–2020. Available online: https://health.gov/dietaryguidelines/2015/guidelines/ (accessed on 12 July 2019).
- Ensembl Plants [Internet]. Available online: https://plants.ensembl.org/index.html (accessed on 22 July 2020).
- Hard Winter Wheat Regional Nursery Program [Internet]. Available online: https://www.ars.usda.gov/plains-area/lincoln-ne/wheat-sorghum-and-forage-research/docs/hard-winter-wheat-regional-nursery-program/research/ (accessed on 22 July 2020).
- Sidhu, J.S.; Singh, D.; Gill, H.S.; Brar, N.K.; Qiu, Y.; Halder, J.; Al Tameemi, R.; Turnipseed, B.; Sehgal, S.K. Genome-Wide Association Study Uncovers Novel Genomic Regions Associated With Coleoptile Length in Hard Winter Wheat. Front. Genet. 2020, 10, 1345. Available online: https://www.frontiersin.org/article/10.3389/fgene.2019.01345/full (accessed on 13 July 2020). [CrossRef]
- GrainGenes [Internet]. Available online: https://wheat.pw.usda.gov/GG3/ (accessed on 22 July 2020).
- Iftikhar, A.; Ali, I. Kernel softness in wheat is determined by starch granule bound Puroindoline proteins. J. Plant Biochem. Biotechnol. 2016, 26, 247–262. [Google Scholar] [CrossRef]
- Pauly, A.; Pareyt, B.; Fierens, E.; Delcour, J.A. Wheat (Triticum aestivum L. and T. turgidum L. ssp. durum) Kernel Hardness: I. Current View on the Role of Puroindolines and Polar Lipids. Compr. Rev. Food Sci. Food Saf. 2013, 12, 413–426. Available online: http://doi.wiley.com/10.1111/1541-4337.12019 (accessed on 13 July 2020).
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; García, B.M.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2011, 35, 454–484. [Google Scholar] [CrossRef]






| Trait | Marker | SNP | Chr. | Position (bp) | p-Value | Marker R2 b | Additive Effect Score (Associated Genotype) | Genotype at SNP Site Associated with an Increase in the Phenotypic Value of the Trait (Number of Lines with This Genotype) a | Genotype at SNP Site Associated with a Decrease in the Phenotypic Value of the Trait (Number of Lines with This Genotype) |
|---|---|---|---|---|---|---|---|---|---|
| Friability | BS00000020_51 | A/G | 5D | 3,609,894 | <0.001 | 0.12 | −5.95 (AA) | GG (46) | AA (250) |
| Excalibur_c49805_63 | A/G | 5D | 1,614,602 | <0.001 | 0.11 | 5.93 (AA) | AA (43) | GG (254) | |
| WRC | IWA4867 | C/T | 4A | 562,407,344 | 0.0002 | 0.05 | −20.5 (CC) | TT (192) | CC (97) |
| IWA4698 | C/T | 4A | 562,453,542 | 0.0002 | 0.04 | −19.9 (CC) | TT (194) | CC (98) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navrotskyi, S.; Belamkar, V.; Baenziger, P.S.; Rose, D.J. Insights into the Genetic Architecture of Bran Friability and Water Retention Capacity, Two Important Traits for Whole Grain End-Use Quality in Winter Wheat. Genes 2020, 11, 838. https://doi.org/10.3390/genes11080838
Navrotskyi S, Belamkar V, Baenziger PS, Rose DJ. Insights into the Genetic Architecture of Bran Friability and Water Retention Capacity, Two Important Traits for Whole Grain End-Use Quality in Winter Wheat. Genes. 2020; 11(8):838. https://doi.org/10.3390/genes11080838
Chicago/Turabian StyleNavrotskyi, Sviatoslav, Vikas Belamkar, P. Stephen Baenziger, and Devin J. Rose. 2020. "Insights into the Genetic Architecture of Bran Friability and Water Retention Capacity, Two Important Traits for Whole Grain End-Use Quality in Winter Wheat" Genes 11, no. 8: 838. https://doi.org/10.3390/genes11080838
APA StyleNavrotskyi, S., Belamkar, V., Baenziger, P. S., & Rose, D. J. (2020). Insights into the Genetic Architecture of Bran Friability and Water Retention Capacity, Two Important Traits for Whole Grain End-Use Quality in Winter Wheat. Genes, 11(8), 838. https://doi.org/10.3390/genes11080838