Plant-Specific Domains and Fragmented Sequences Imply Non-Canonical Functions in Plant Aminoacyl-tRNA Synthetases
Abstract
:1. Introduction
2. Materials and Methods
Bioinformatics
3. Results
3.1. aaRS Genes in Arabidopsis
3.2. Fragmented Sequences of aaRSs Found in the Arabidopsis Genome
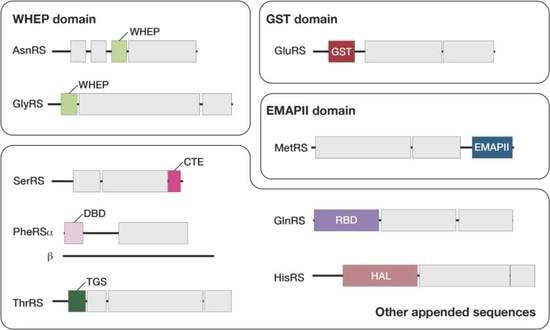
3.3. Plant-Specific Domains Found in Arabidopsis aaRSs
3.4. Plant-Specific WHEP Domain Insertion in AsnRS
3.5. Plant-Specific Long N-Terminal Extension in HisRS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guo, M.; Yang, X.L.; Schimmel, P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 2010, 11, 668–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sajish, M.; Zhou, Q.; Kishi, S.; Valdez, D.M., Jr.; Kapoor, M.; Guo, M.; Lee, S.; Kim, S.; Yang, X.L.; Schimmel, P. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-γ and p53 signaling. Nat. Chem. Biol. 2012, 8, 547–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.M.; Jeong, S.J.; Park, M.C.; Kim, G.; Kwon, N.H.; Kim, H.K.; Ha, S.H.; Ryu, S.H.; Kim, S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012, 149, 410–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Kapoor, M.; Guo, M.; Belani, R.; Xu, X.; Kiosses, W.B.; Hanan, M.; Park, C.; Armour, E.; Do, M.H.; et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat. Struct. Mol. Biol. 2010, 17, 57–61. [Google Scholar] [CrossRef] [Green Version]
- Sajish, M.; Schimmel, P. A human tRNA synthetase is a potent PARP1-activating effector target for resveratrol. Nature 2015, 519, 370–373. [Google Scholar] [CrossRef] [Green Version]
- Lo, W.S.; Gardiner, E.; Xu, Z.; Lau, C.F.; Wang, F.; Zhou, J.J.; Mendlein, J.D.; Nangle, L.A.; Chiang, K.P.; Yang, X.L.; et al. Human tRNA synthetase catalytic nulls with diverse functions. Science 2014, 345, 328–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, C.A.; Kuhla, B.; Cusack, S.; Lambowitz, A.M. tRNA-like recognition of group I introns by a tyrosyl-tRNA synthetase. Proc. Natl. Acad. Sci. USA 2002, 99, 2630–2635. [Google Scholar] [CrossRef] [Green Version]
- Frechin, M.; Enkler, L.; Tetaud, E.; Laporte, D.; Senger, B.; Blancard, C.; Hammann, P.; Bader, G.; Clauder-Münster, S.; Steinmetz, L.M.; et al. Expression of nuclear and mitochondrial genes encoding ATP synthase is synchronized by disassembly of a multisynthetase complex. Mol. Cell 2014, 56, 763–776. [Google Scholar] [CrossRef] [Green Version]
- Luna, E.; van Hulten, M.; Zhang, Y.; Berkowitz, O.; López, A.; Pétriacq, P.; Sellwood, M.A.; Chen, B.; Burrell, M.; van de Meene, A.; et al. Plant perception of aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nat. Chem. Biol. 2014, 10, 450–456. [Google Scholar] [CrossRef] [Green Version]
- Kekez, M.; Zanki, V.; Kekez, I.; Baranasic, J.; Hodnik, V.; Duchene, A.M.; Anderluh, G.; Gruic-Sovulj, I.; Matkovic-Calogovic, D.; Weygand-Durasevic, I.; et al. Arabidopsis seryl-tRNA synthetase: The first crystal structure and novel protein interactor of plant aminoacyl-tRNA synthetase. FEBS J. 2019, 286, 536–554. [Google Scholar] [CrossRef] [Green Version]
- Zuo, D.Y.; Yi, S.Y.; Liu, R.J.; Qu, B.; Huang, T.; He, W.J.; Li, C.; Li, H.P.; Liao, Y.C. A deoxynivalenol-activated methionyl-tRNA synthetase gene from wheat encodes a nuclear localized protein and protects plants against Fusarium pathogens and mycotoxins. Phytopathology 2016, 106, 614–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uwer, U.; Willmitzer, L.; Altmann, T. Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell 1998, 10, 1277–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moschopoulos, A.; Derbyshire, P.; Byrne, M.E. The Arabidopsis organelle-localized glycyl-tRNA synthetase encoded by EMBRYO DEFECTIVE DEVELOPMENT1 is required for organ patterning. J. Exp. Bot. 2012, 63, 5233–5243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giritch, A.; Herbik, A.; Balzer, H.J.; Ganal, M.; Stephan, U.W.; Bäumlein, H. A root-specific iron-regulated gene of tomato encodes a lysyl-tRNA-synthetase-like protein. Eur. J. Biochem. 1997, 244, 310–317. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Somerville, C. Suspensor-derived polyembryony caused by altered expression of valyl-tRNA synthetase in the twn2 mutant of Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7349–7355. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Wang, S.; Li, H.; Zhang, G.; Li, H. EMBRYONIC FACTOR 31 encodes a tyrosyl-tRNA synthetase that is essential for seed development. Mol. Biol. Rep. 2012, 39, 8297–8305. [Google Scholar] [CrossRef]
- Yang, X.; Li, G.; Tian, Y.; Song, Y.; Liang, W.; Zhang, D. A rice glutamyl-tRNA synthetase modulates early anther cell division and patterning. Plant Physiol. 2018, 177, 728–744. [Google Scholar] [CrossRef] [Green Version]
- Duchene, A.M.; Giritch, A.; Hoffmann, B.; Cognat, V.; Lancelin, D.; Peeters, N.M.; Zaepfel, M.; Maréchal-Drouard, L.; Small, I.D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 16484–16489. [Google Scholar] [CrossRef] [Green Version]
- Larson, E.T.; Kim, J.E.; Castaneda, L.J.; Napuli, A.J.; Zhang, Z.; Fan, E.; Zucker, F.H.; Verlinde, C.L.M.J.; Buckner, F.S.; Van Voorhis, W.C.; et al. The double-length tyrosyl-tRNA synthetase from the eukaryote Leishmania major forms an intrinsically asymmetric pseudo-dimer. J. Mol. Biol. 2011, 409, 159–176. [Google Scholar] [CrossRef] [Green Version]
- Barros-Álvarez, X.; Kerchner, K.M.; Koh, C.Y.; Turley, S.; Pardon, E.; Steyaert, J.; Ranade, R.M.; Gillespie, J.R.; Zhang, Z.; Verlinde, C.L.M.J.; et al. Leishmania donovani tyrosyl-tRNA synthetase structure in complex with a tyrosyl adenylate analog and comparisons with human and protozoan counterparts. Biochimie 2017, 138, 124–136. [Google Scholar] [CrossRef] [Green Version]
- Pujol, C.; Bailly, M.; Kern, D.; Maréchal-Drouard, L.; Becker, H.; Duchene, A.M. Dual-targeted tRNA-dependent amidotransferase ensures both mitochondrial and chloroplastic Gln-tRNAGln synthesis in plants. Proc. Natl. Acad. Sci. USA 2008, 105, 6481–6485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, M.; Schimmel, P.; Yang, X.L. Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 2010, 584, 434–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaminska, M.; Deniziak, M.; Kerjan, P.; Barciszewski, J.; Mirande, M. A recurrent general RNA binding domain appended to plant methionyl-tRNA synthetase acts as a cis-acting cofactor for aminoacylation. EMBO J. 1999, 24, 6908–6917. [Google Scholar] [CrossRef] [PubMed]
- Peeters, N.M.; Chapron, A.; Giritch, A.; Grandjean, O.; Lancelin, D.; Lhomme, T.; Vivrel, A.; Small, I. Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J. Mol. Evol. 2000, 50, 413–423. [Google Scholar] [CrossRef] [Green Version]
- Pilbák, S.; Tomin, A.; Rétey, J.; Poppe, L. The essential tyrosine-containing loop conformation and the role of the C-terminal multi-helix region in eukaryotic phenylalanine ammonia-lyases. FEBS J. 2006, 273, 1004–1019. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.F.; Rétry, J.; Schulz, G.E. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 1999, 38, 5355–5361. [Google Scholar] [CrossRef]
- Ahel, I.; Korencic, D.; Ibba, M.; Söll, D. Trans-editing of mischarged tRNAs. Proc. Natl. Acad. Sci. USA 2003, 100, 15422–15427. [Google Scholar] [CrossRef] [Green Version]
- Vargas-Rodriguez, O.; Musier-Forsyth, K. Exclusive use of trans-editing domains prevents proline mistranslation. J. Biol. Chem. 2013, 288, 14391–14399. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Vargas-Rodriguez, O.; Goto, Y.; Novoa, E.M.; de Pouplana, L.R.; Suga, H.; Musier-Forsyth, K. Homologous trans-editing factors with broad tRNA specificity prevent mistranslation caused by serine/threonine misactivation. Proc. Natl. Acad. Sci. USA 2015, 112, 6027–6032. [Google Scholar] [CrossRef] [Green Version]
- Korencic, D.; Ahel, I.; Schelert, J.; Sacher, M.; Ruan, B.; Stathopoulos, C.; Blum, P.; Ibba, M.; Söll, D. A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc. Natl. Acad. Sci. USA 2004, 101, 10260–10265. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Hale, S.P.; Schimmel, P. Aminoacylation error correction. Nature 1996, 384, 33–34. [Google Scholar] [CrossRef] [PubMed]
- Nureki, O.; Vassylyev, D.G.; Tateno, M.; Shimada, A.; Nakama, T.; Fukai, S.; Konno, M.; Hendrickson, T.L.; Schimmel, P.; Yokoyama, S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science 1998, 280, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Champagne, K.S.; Sissler, M.; Larrabee, Y.; Doublié, S.; Francklyn, C.S. Activation of the hetero-octameric ATP phosphoribosyl transferase through subunit interface rearrangement by a tRNA synthetase paralog. J. Biol. Chem. 2005, 280, 34096–34104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sissler, M.; Delorme, C.; Bond, J.; Ehrlich, S.D.; Renault, P.; Francklyn, C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 8985–8990. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Class I | AGI No. | Localization Prediction | Primary Structure | Class II | AGI No. | Localization Prediction | Primary Structure |
|---|---|---|---|---|---|---|---|
| ArgRS | At4g26300 | cytosol, cp | AlaRS | At1g50200 | cytosol, mt | ||
| At1g66530 | cytosol, mt | At5g22800 | mt, cp | ||||
| CysRS | At5g38830 | cytosol | At1g49930 | N truncated | |||
| At3g56300 | cytosol | AsnRS | At5g56680 | cytosol | |||
| At2g31170 | mt, cp | At4g17300 | mt, cp | ||||
| GlnRS | At1g25350 | cytosol | At1g70980 | cytosol | |||
| At5g19720 | N truncated | At3g07420 | cytosol | ||||
| GluRS | At5g26710 | cytosol | At1g68420 | N, C truncated | |||
| At5g64050 | mt, cp | At5g38750 | N, C truncated | ||||
| IleRS | At5g49030 | mt, cp | AspRS | At4g33760 | mt, cp | ||
| At4g10320 | cytosol | At4g31180 | cytosol | ||||
| At3g23145 | N, C truncated | At4g26870 | cytosol | ||||
| LeuRS | At4g04350 | cp | GlyRS | At3g48110 | mt, cp | ||
| At1g09620 | cytosol, mt | At1g29880 | cytosol, mt | ||||
| MetRS | At4g13780 | cytosol | At3g44740 | N, C truncated | |||
| At3g55400 | mt, cp | At1g29870 | C truncated | ||||
| At5g02680 | N, C truncated | HisRS | At3g02760 | cytosol | |||
| TrpRS | At3g04600 | cytosol | At3g46100 | mt, cp | |||
| At2g25840 | mt, cp | At5g03406 | C truncated | ||||
| TyrRS | At3g02660 | mt, cp | LysRS | At3g11710 | cytosol | ||
| At2g33840 | cytosol | At3g13490 | mt, cp | ||||
| At1g28350 | cytosol | pseudo-dimer | At3g30805 | N truncated | |||
| ValRS | At5g16715 | mt, cp | PheRS | At4g39280 | cytosol | ||
| At1g14610 | cytosol, mt | At1g72550 | cytosol | ||||
| At1g27160 | N truncated | At3g58140 | mt, cp | ||||
| ProRS | At5g52520 | mt, cp | |||||
| At3g62120 | cytosol | ||||||
| At5g10880 | N truncated | ||||||
| SerRS | At5g27470 | cytosol | |||||
| At1g11870 | mt, cp | ||||||
| ThrRS | At5g26830 | cytosol, mt | |||||
| At2g04842 | mt, cp | ||||||
| At1g18130 | N truncated | ||||||
| At1g17960 | motif1 missing |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saga, Y.; Kawashima, M.; Sakai, S.; Yamazaki, K.; Kaneko, M.; Takahashi, M.; Sato, N.; Toyoda, Y.; Takase, S.; Nakano, T.; et al. Plant-Specific Domains and Fragmented Sequences Imply Non-Canonical Functions in Plant Aminoacyl-tRNA Synthetases. Genes 2020, 11, 1056. https://doi.org/10.3390/genes11091056
Saga Y, Kawashima M, Sakai S, Yamazaki K, Kaneko M, Takahashi M, Sato N, Toyoda Y, Takase S, Nakano T, et al. Plant-Specific Domains and Fragmented Sequences Imply Non-Canonical Functions in Plant Aminoacyl-tRNA Synthetases. Genes. 2020; 11(9):1056. https://doi.org/10.3390/genes11091056
Chicago/Turabian StyleSaga, Yusuke, Moeka Kawashima, Shiho Sakai, Kaori Yamazaki, Misato Kaneko, Moeka Takahashi, Natsuko Sato, Yohei Toyoda, Shohei Takase, Takeshi Nakano, and et al. 2020. "Plant-Specific Domains and Fragmented Sequences Imply Non-Canonical Functions in Plant Aminoacyl-tRNA Synthetases" Genes 11, no. 9: 1056. https://doi.org/10.3390/genes11091056
APA StyleSaga, Y., Kawashima, M., Sakai, S., Yamazaki, K., Kaneko, M., Takahashi, M., Sato, N., Toyoda, Y., Takase, S., Nakano, T., Kawakami, N., & Kushiro, T. (2020). Plant-Specific Domains and Fragmented Sequences Imply Non-Canonical Functions in Plant Aminoacyl-tRNA Synthetases. Genes, 11(9), 1056. https://doi.org/10.3390/genes11091056