APOE Variants in an Iberian Alzheimer Cohort Detected through an Optimized Sanger Sequencing Protocol
Abstract
:1. Introduction
2. Materials and Methods
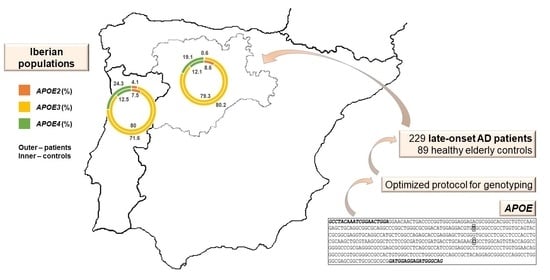
2.1. Iberian Cohort
2.2. Sample Collection and DNA Extraction
2.3. Whole Genome Amplification
2.4. APOE Genotyping
2.5. Sequencing Results and Statistical Analysis
3. Results
3.1. Optimization of APOE Genotyping
3.2. APOE Genetic Variation and LOAD ORs in Northern Portugal and in Castile and León Region, Spain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alzheimer’s Association. 2008 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2008, 4, 110–133. [Google Scholar] [CrossRef]
- Schachter, A.S.; Davis, K.L. Alzheimer’s Disease. Dialogues Clin. Neurosci. 2000, 2, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gleerup, H.S.; Hasselbalch, S.G.; Simonsen, A.H. Biomarkers for Alzheimer’s Disease in Saliva: A Systematic Review. Dis. Markers 2019, 2019, 4761054. [Google Scholar] [CrossRef] [PubMed]
- Joe, E.; Ringman, J.M. Cognitive symptoms of Alzheimer’s disease: Clinical management and prevention. BMJ 2019, 367, l6217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloom, G.S. Amyloid-β and Tau. JAMA Neurol. 2014, 71, 505–508. [Google Scholar] [CrossRef] [Green Version]
- Reale, M.; Gonzales-Portillo, I.; Borlongan, C.V. Saliva, an easily accessible fluid as diagnostic tool and potent stem cell source for Alzheimer’s Disease: Present and future applications. Brain Res. 2020, 1727, 146535. [Google Scholar] [CrossRef]
- Blennow, K.; Zetterberg, H. Biomarkers for Alzheimer’s disease: Current status and prospects for the future. J. Intern. Med. 2018, 284, 643–663. [Google Scholar] [CrossRef] [Green Version]
- Buschmann, D.; González, R.; Kirchner, B.; Mazzone, C.; Pfaffl, M.W.; Schelling, G.; Steinlein, O.; Reithmair, M. Glucocorticoid receptor overexpression slightly shifts microRNA expression patterns in triple-negative breast cancer. Int. J. Oncol. 2018, 52, 1765–1776. [Google Scholar] [CrossRef]
- Ross, C.A.; Aylward, E.H.; Wild, E.J.; Langbehn, D.R.; Long, J.D.; Warner, J.H.; Scahill, R.I.; Leavitt, B.R.; Stout, J.C.; Paulsen, J.S.; et al. Huntington disease: Natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol. 2014, 10, 204–216. [Google Scholar] [CrossRef] [Green Version]
- Frieden, C.; Garai, K. Concerning the structure of apoE. Protein Sci. 2013, 22, 1820–1825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kushioka, T.; Ocho, M.; Ito, Y.; Yokokawa, T.; Yui, K.; Yamasaki, Y. Evaluation of ApoE Genotyping Using Saliva-Derived DNA. J. Clin. Med Genom. 2018, 6, 2. [Google Scholar] [CrossRef]
- Heffernan, A.L.; Chidgey, C.; Peng, P.; Masters, C.L.; Roberts, B.R. The Neurobiology and Age-Related Prevalence of the ε4 Allele of Apolipoprotein E in Alzheimer’s Disease Cohorts. J. Mol. Neurosci. 2016, 60, 316–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, G.C.; Insel, P.; Tosun, D.; Schuff, N.; Truransacrey, D.; Raptentsetsang, S.T.; Jack, C.R.; Aisen, P.S.; Petersen, R.C.; Weiner, M.W.; et al. Hippocampal atrophy rates and CSF biomarkers in elderly APOE2 normal subjects. Neurology 2010, 75, 1976–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belloy, M.E.; Napolioni, V.; Greicius, M.D. A Quarter Century of APOE and Alzheimer’s Disease: Progress to Date and the Path Forward. Neuron 2019, 101, 820–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahley, R.W. Apolipoprotein E: From cardiovascular disease to neurodegenerative disorders. J. Mol. Med. 2016, 94, 739–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muza, P.; Bachmeier, C.; Mouzon, B.; Algamal, M.; Rafi, N.G.; Lungmus, C.; Abdullah, L.; Evans, J.E.; Ferguson, S.; Mullan, M.; et al. APOE Genotype Specific Effects on the Early Neurodegenerative Sequelae Following Chronic Repeated Mild Traumatic Brain Injury. Neuroscience 2019, 404, 297–313. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Borgström, E.; Paterlini, M.; Mold, J.E.; Frisen, J.; Lundeberg, J. Comparison of whole genome amplification techniques for human single cell exome sequencing. PLoS ONE 2017, 12, e0171566. [Google Scholar] [CrossRef] [Green Version]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, A.; Korf, B. Genetic Testing Techniques. In Pediatric Cancer Genetics; Elsevier BV: Amsterdam, The Netherlands, 2018; pp. 47–64. [Google Scholar]
- Hagemann, I.S. Overview of Technical Aspects and Chemistries of Next-Generation Sequencing. In Clinical Genomics; Elsevier BV: Amsterdam, The Netherlands, 2015; pp. 3–19. [Google Scholar]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Korbie, D.J.; Mattick, J.S. Touchdown PCR for Increased Specificity and Sensitivity in PCR Amplification. Nat. Protoc. 2008, 3, 1452–1456. [Google Scholar] [CrossRef] [PubMed]
- Haddy, N.; De Bacquer, D.; Chemaly, M.M.; Maurice, M.; Ehnholm, C.; Evans, A.; Sans, S.; Martins, M.D.C.; De Backer, G.; Siest, G.; et al. The importance of plasma apolipoprotein E concentration in addition to its common polymorphism on inter-individual variation in lipid levels: Results from Apo Europe. Eur. J. Hum. Genet. 2002, 10, 841–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seixas, S.; Trovoada, M.J.; Rocha, J. Haplotype analysis of the apolipoprotein E and apolipoprotein C1 loci in Portugal and São Tomé e Príncipe (Gulf of Guinea): Linkage disequilibrium evidence that APOE*4 is the ancestral APOE allele. Hum. Biol. 1999, 71, 1001–1008. [Google Scholar] [PubMed]
- Ibarreta, L.; Gómez-Isla, T.; Portera-Sanchez, A.; Parrilla, R.; Ayuso-Parrilla, M.S. Apolipoprotein E genotype in Spanish patients of Alzheimer’s or Parkinson’s disease. J. Neurol. Sci. 1995, 134, 146–149. [Google Scholar] [CrossRef]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; van Duijn, C.M. Effects of Age, Sex, and Ethnicity on the Association between Apolipoprotein E Genotype and Alzheimer Disease. A Meta-Analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef]
- Corbo, R.M.; Scacchi, R. Apolipoprotein E (APOE) allele distribution in the world. IsAPOE*4a ‘thrifty’ allele? Ann. Hum. Genet. 1999, 63, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.P.; Singh, M.; Mastana, S. APOE distribution in world populations with new data from India and the UK. Ann. Hum. Biol. 2006, 33, 279–308. [Google Scholar] [CrossRef] [PubMed]
- Egert, S.; Rimbach, G.; Huebbe, P. ApoE genotype: From geographic distribution to function and responsiveness to dietary factors. Proc. Nutr. Soc. 2012, 71, 410–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortensen, E.L.; Høgh, P. A gender difference in the association between APOE genotype and age-related cognitive decline. Neurology 2001, 57, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Tudorache, I.F.; Trusca, V.G.; Gafencu, A.V. Apolipoprotein E-a Multifunctional Protein with Implications in Various Pathologies as a Result of Its Structural Features. Comput. Struct. Biotechnol. J. 2017, 15, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Toro, C.A.; Zhang, L.; Cao, J.; Cai, D. Sex differences in Alzheimer’s disease: Understanding the molecular impact. Brain Res. 2019, 1719, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Neu, S.C.; Pa, J.; Kukull, W.; Beekly, D.; Kuzma, A.; Gangadharan, P.; Wang, L.-S.; Romero, K.; Arneric, S.P.; Redolfi, A.; et al. Apolipoprotein E Genotype and Sex Risk Factors for Alzheimer Disease. JAMA Neurol. 2017, 74, 1178–1189. [Google Scholar] [CrossRef]
- Nebel, R.A.; Aggarwal, N.T.; Barnes, L.L.; Gallagher, A.; Goldstein, J.M.; Kantarci, K.; Mallampalli, M.P.; Mormino, E.C.; Scott, L.; Yu, W.H.; et al. Understanding the impact of sex and gender in Alzheimer’s disease: A call to action. Alzheimer’s Dement. 2018, 14, 1171–1183. [Google Scholar] [CrossRef]


| Controls | LOAD Patients | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| MIL | MOD | SEV | |||||||
| Population | PT | SP | PT | SP | PT | SP | PT | SP | |
| Females (N) | 37 | 13 | 20 | 14 | 27 | 21 | 66 | 22 | |
| Males (N) | 23 | 16 | 7 | 14 | 8 | 5 | 20 | 5 | |
| Examination age (years) | Range | 70–96 | 68–87 | 65–93 * | 62–89 | 67–96 | 63–92 | 65–99 | 66–94 |
| Mean ± SD | 84.3 ± 7.3 | 77.3 ± 4.8 | 80.2 ± 7.6 | 78.1 ± 6.9 | 83.4 ± 8.1 | 79.9 ± 7.6 | 83.4 ± 7.8 | 82.8 ± 7.0 | |
| MMSE | Range | 26–30 | 27–30 | 12–24 ** | 20–30 | 5–28 ** | 11–20 | 0–10 | 0–10 |
| Mean ± SD | 28.2 ± 1.2 | 29.1 ± 1.1 | 20.0 ± 3.6 | 23.2 ± 2.5 | 12.0 ± 4.6 | 14.7 ± 2.7 | 1.3 ± 2.9 | 4.4 ± 3.9 | |
| Status | Population | Source | N | APOE Allele Frequency (%) | APOE Genotype Frequency (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E2 | E3 | E4 | E2/E2 | E2/E3 | E2/E4 | E3/E3 | E3/E4 | E4/E4 | ||||
| Controls | Portuguese | This study | 60 | 7.5 | 80.0 | 12.5 | 1.7 | 11.7 | 0 | 66.7 | 15.0 | 5.0 |
| Portuguese | Haddy et al., 2002 [28] | 607 § | 6.3 | 84.3 | 9.4 | 0.3 | 11.0 | 1.0 | 71.2 | 15.2 | 1.3 | |
| Portuguese | Seixas et al., 1999 [29] | 149 ʄ | 4.4 | 88.2 | 7.4 | 0 | 8.7 | 0 | 77.9 | 12.1 | 1.3 | |
| Spanish | This study | 29 | 8.6 | 79.3 | 12.1 | 0 | 17.2 | 0 | 65.5 | 10.3 | 6.9 | |
| Spanish | Ibarreta et al., 1995 [30] | 42 # | 5.0 | 92.0 | 4.0 | NA | NA | NA | NA | NA | NA | |
| Spanish | Haddy et al., 2002 [28] | 906 §§ | 7.8 | 81.1 | 11.1 | 0.9 | 12.7 | 1.1 | 64.9 | 19.6 | 0.8 | |
| Caucasian *** | Farrer et al., 1997 [31] | 6262 * | 8.4 | 77.9 | 13.7 | 0.8 | 12.7 | 2.6 | 60.9 | 21.3 | 1.8 | |
| Patients | Portuguese | This study | 148 | 4.1 | 71.6 | 24.3 | 1.4 | 5.4 | 0 | 52.7 | 32.4 | 8.1 |
| Spanish | This study | 81 | 0.6 | 80.2 | 19.1 | 0 | 1.2 | 0 | 64.2 | 30.9 | 3.7 | |
| Spanish | Ibarreta et al., 1995 [30] | 47## | 6.0 | 60.0 | 34.0 | NA | NA | NA | NA | NA | NA | |
| Caucasian *** | Farrer et al., 1997 [31] | 5107 ** | 3.9 | 59.4 | 36.7 | 0.2 | 4.8 | 2.6 | 36.4 | 41.1 | 14.8 | |
| Portugal | Spain | ||||
|---|---|---|---|---|---|
| LOAD Patients | Controls | LOAD Patients | Controls | ||
| Allele Distribution | |||||
| Portugal | LOAD patients (N = 296) | 0.00887 ± 0.00173 * | 0.03265 ± 0.00504 * | NC | |
| Controls (N = 120) | NC | 0.95815 ± 0.00102 | |||
| Spain | LOAD patients (N = 162) | 0.00560 ± 0.00118 * | |||
| Controls (N = 58) | |||||
| Genotype Distribution | |||||
| Portugal | LOAD patients (N = 148) | 0.03801 ± 0.00308 * | NC | NC | |
| Spain | LOAD patients (N = 81) | NC | NC | 0.00232 ± 0.00051 * | |
| Population | APOE Genotype/Allele | Controls (N) | Patients (N) | Odds Ratio (95% Confidence Interval) | p-Value |
|---|---|---|---|---|---|
| Portuguese (this study) | E3E3 (Referent) | 40 | 78 | 1 | - |
| E2E2 | 1 | 2 | 1.03 (0.09 to 11.66) | 0.9837 | |
| E2E3 | 7 | 8 | 0.59 (0.20 to 1.73) | 0.3338 | |
| E2E4 | 0 | 0 | - | - | |
| E3E4 | 9 | 48 | 2.74 (1.22 to 6.13) | 0.0146 | |
| E4E4 | 3 | 12 | 2.05 (0.55 to 7.69) | 0.2865 | |
| E3 (Referent) | 96 | 212 | 1 | - | |
| E2 | 9 | 12 | 0.60 (0.25 to 1.48) | 0.2704 | |
| E4 | 15 | 72 | 2.17 (1.19 to 3.99) | 0.0121 | |
| Spanish (this study) | E3E3 (Referent) | 19 | 52 | 1 | - |
| E2E2 | 0 | 0 | - | - | |
| E2E3 | 5 | 1 | 0.07 (0.01 to 0.67) | 0.0203 | |
| E2E4 | 0 | 0 | - | - | |
| E3E4 | 3 | 25 | 3.04 (0.82 to 11.26) | 0.0952 | |
| E4E4 | 2 | 3 | 0.55 (0.08 to 3.54) | 0.5274 | |
| E3 (Referent) | 46 | 130 | 1 | - | |
| E2 | 5 | 1 | 0.07 (0.01 to 0.62) | 0.0169 | |
| E4 | 7 | 31 | 1.57 (0.65 to 3.80) | 0.3206 | |
| Caucasian (Farrer et al., 1997) | E3E3 (Referent) | 3813 | 1859 | 1 | - |
| E2E2 | 50 | 10 | 0.41 (0.21 to 0.81) | 0.0104 | |
| E2E3 | 795 | 245 | 0.63 (0.54 to 0.74) | <0.0001 | |
| E2E4 | 163 | 133 | 1.67 (1.32 to 2.12) | <0.0001 | |
| E3E4 | 1328 | 2104 | 3.25 (2.97 to 3.55) | <0.0001 | |
| E4E4 | 113 | 756 | 13.72 (11.18 to 16.85) | <0.0001 | |
| E3 (Referent) | 9756 | 6067 | 1 | - | |
| E2 | 1052 | 398 | 0.61 (0.54 to 0.69) | <0.0001 | |
| E4 | 1716 | 3749 | 3.51 (3.29 to 3.75) | <0.0001 |
| Portuguese Cohort | Spanish Cohort | ||||||
|---|---|---|---|---|---|---|---|
| Total | Females | Males | Total | Females | Males | ||
| Patients (N) | APOE4 carriers | 60 | 46 | 14 | 28 | 20 | 8 |
| APOE4 non-carriers | 88 | 67 | 21 | 53 | 37 | 16 | |
| Controls (N) | APOE4 carriers | 12 | 8 | 4 | 5 | 1 | 4 |
| APOE4 non-carriers | 48 | 29 | 19 | 24 | 12 | 12 | |
| Odds ratio (95% confidence interval) | 2.73 (1.34 to 5.56) | 2.49 (1.04 to 5.93) | 3.17 (0.89 to 11.31) | 2.54 (0.87 to 7.37) | 6.49 (0.79 to 53.57) | 1.50 (0.36 to 6.17) | |
| p-value | 0.0058 | 0.0395 | 0.0759 | 0.0873 | 0.0826 | 0.5742 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González, R.D.; Gomes, I.; Gomes, C.; Rocha, R.; Durães, L.; Sousa, P.; Figueruelo, M.; Rodríguez, M.; Pita, C.; Hornero, R.; et al. APOE Variants in an Iberian Alzheimer Cohort Detected through an Optimized Sanger Sequencing Protocol. Genes 2021, 12, 4. https://doi.org/10.3390/genes12010004
González RD, Gomes I, Gomes C, Rocha R, Durães L, Sousa P, Figueruelo M, Rodríguez M, Pita C, Hornero R, et al. APOE Variants in an Iberian Alzheimer Cohort Detected through an Optimized Sanger Sequencing Protocol. Genes. 2021; 12(1):4. https://doi.org/10.3390/genes12010004
Chicago/Turabian StyleGonzález, Ricardo D., Iva Gomes, Catarina Gomes, Rita Rocha, Luís Durães, Patrícia Sousa, Manuel Figueruelo, Maria Rodríguez, Carmen Pita, Roberto Hornero, and et al. 2021. "APOE Variants in an Iberian Alzheimer Cohort Detected through an Optimized Sanger Sequencing Protocol" Genes 12, no. 1: 4. https://doi.org/10.3390/genes12010004
APA StyleGonzález, R. D., Gomes, I., Gomes, C., Rocha, R., Durães, L., Sousa, P., Figueruelo, M., Rodríguez, M., Pita, C., Hornero, R., Gómez, C., Lopes, A. M., Pinto, N., & Martins, S. (2021). APOE Variants in an Iberian Alzheimer Cohort Detected through an Optimized Sanger Sequencing Protocol. Genes, 12(1), 4. https://doi.org/10.3390/genes12010004